mKikGR, a monomeric photoswitchable fluorescent protein
- PMID: 19079591
- PMCID: PMC2592542
- DOI: 10.1371/journal.pone.0003944
mKikGR, a monomeric photoswitchable fluorescent protein
Abstract
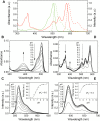
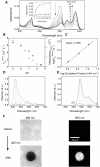
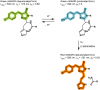
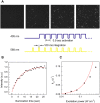
The recent demonstration and utilization of fluorescent proteins whose fluorescence can be switched on and off has greatly expanded the toolkit of molecular and cell biology. These photoswitchable proteins have facilitated the characterization of specifically tagged molecular species in the cell and have enabled fluorescence imaging of intracellular structures with a resolution far below the classical diffraction limit of light. Applications are limited, however, by the fast photobleaching, slow photoswitching, and oligomerization typical for photoswitchable proteins currently available. Here, we report the molecular cloning and spectroscopic characterization of mKikGR, a monomeric version of the previously reported KikGR that displays high photostability and switching rates. Furthermore, we present single-molecule imaging experiments that demonstrate that individual mKikGR proteins can be localized with a precision of better than 10 nanometers, suggesting their suitability for super-resolution imaging.
Conflict of interest statement
Figures


References
-
- Tsien RY. The green fluorescent protein. Annual Review of Biochemistry. 1998;67:509–544. - PubMed
-
- Zhang J, Campbell RE, Ting AY, Tsien RY. Creating new fluorescent probes for cell biology. Nature Reviews Molecular Cell Biology. 2002;3:906–918. - PubMed
-
- Miyawaki A, Sawano A, Kogure T. Lighting up cells: labelling proteins with fluorophores. Nature Cell Biology. 2003:S1–S7. - PubMed
-
- Chudakov DM, Lukyanov S, Lukyanov KA. Fluorescent proteins as a toolkit for in vivo imaging. Trends in Biotechnology. 2005;23:605–613. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

