Polymersome delivery of siRNA and antisense oligonucleotides
- PMID: 19084037
- PMCID: PMC2740336
- DOI: 10.1016/j.jconrel.2008.10.020
Polymersome delivery of siRNA and antisense oligonucleotides
Erratum in
- J Control Release. 2009 Aug 4;137(3):256. Williams, Jason [corrected to Williams, Jason H]; Sirsi, Shashank, [corrected to Sirsi, Shashank R]; Lutz, Gordon [corrected to Lutz, Gordon J]
Abstract
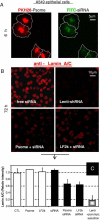
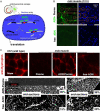
siRNA and antisense oligonucleotides, AON, have similar size and negative charge and are often packaged for in vitro delivery with cationic lipids or polymers-but exposed positive charge is problematic in vivo. Here we demonstrate loading and functional delivery of RNAi and AON with non-ionic, nano-transforming polymersomes. These degradable carriers are taken up passively by cultured cells after which the vesicles transform into micelles that allow endolysosomal escape and delivery of either siRNA into cytosol for mRNA knockdown or else AON into the nucleus for exon skipping within pre-mRNA. Polymersome-mediated knockdown appears as efficient as common cationic-lipid transfection and about half as effective as Lenti-virus after sustained selection. For AON, initial results also show that intramuscular injection into a mouse model of muscular dystrophy leads to the expected protein expression, which occurs along the entire length of muscle. The lack of cationic groups in antisense polymersomes together with initial tests of efficacy suggests broader utility of these non-viral carriers.
Figures



References
-
- Dillon CP, Sandy P, Nencioni A, Kissler S, Rubinson DA, Van Parijs L. Rnai as an experimental and therapeutic tool to study and regulate physiological and disease processes. Annual Review of Physiology. 2005;67(1):147–173. - PubMed
-
- Fire A, Xu S, Montgomery MK, Kostas SA, Driver SE, Mello CC. Potent and specific genetic interference by double-stranded RNA in Caenorhabditis elegans. Nature. 1998;391:806–811. - PubMed
-
- Lavrovsky Y, Chen S, Roy AK. Therapeutic potential and mechanism of action of oligonucleotides and ribozymes. Biochemical and Molecular Medicine. 1997;62(1):11–22. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

