Mesenchymal origin of hepatic stellate cells, submesothelial cells, and perivascular mesenchymal cells during mouse liver development
- PMID: 19085956
- PMCID: PMC2673117
- DOI: 10.1002/hep.22721
Mesenchymal origin of hepatic stellate cells, submesothelial cells, and perivascular mesenchymal cells during mouse liver development
Abstract
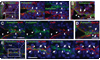
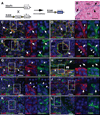
The knowledge concerning fetal hepatic stellate cells (HSCs) is scarce, and their cell lineage and functions are largely unknown. The current study isolated fetal liver mesenchymal cells from a mouse expressing beta-galactosidase under the control of Msx2 promoter by fluorescence-activated cell sorting (FACS) and surveyed marker genes by microarray analysis. Based on the location and immunostaining with conventional and newly disclosed markers, we have identified three distinct populations of fetal liver mesenchymal cells expressing both desmin and p75 neurotrophin receptor (p75NTR): HSCs in the liver parenchyma; perivascular mesenchymal cells expressing alpha-smooth muscle actin (alpha-SMA); and submesothelial cells associated with the basal lamina beneath mesothelial cells and expressing activated leukocyte cell adhesion molecule (ALCAM) and platelet-derived growth factor receptor alpha. A transitional cell type from the submesothelial cell phenotype to fetal HSCs was also identified near the liver surface. Mesothelial cells expressed podoplanin and ALCAM. Ki-67 staining showed that proliferative activity of the submesothelial cells is higher than that of mesothelial cells and transitional cells. Using anti-ALCAM antibodies, submesothelial and mesothelial cells were isolated by FACS. The ALCAM(+) cells expressed hepatocyte growth factor and pleiotrophin. In culture, the ALCAM(+) cells rapidly acquired myofibroblastic morphology and alpha-SMA expression. The ALCAM(+) cells formed intracellular lipid droplets when embedded in collagen gel and treated with retinol, suggesting the potential for ALCAM(+) cells to differentiate to HSCs. Finally, we demonstrated that fetal HSCs, submesothelial cells, and perivascular mesenchymal cells are all derived from mesoderm by using MesP1-Cre and ROSA26 reporter mice.
Conclusion: Fetal HSCs, submesothelial cells, and perivascular mesenchymal cells are mesodermal in origin, and ALCAM(+) submesothelial cells may be a precursor for HSCs in developing liver.
Conflict of interest statement
Potential conflict of interest: Nothing to report.
Figures






Comment in
-
Mesenchymal origin of hepatic stellate cells, submesothelial cells, and perivascular mesenchymal cells during mouse liver development.Hepatology. 2009 Jul;50(1):320; author reply 320. doi: 10.1002/hep.23003. Hepatology. 2009. PMID: 19554539 No abstract available.
References
-
- Geerts A. History, heterogeneity, developmental biology, and functions of quiescent hepatic stellate cells. Semin Liver Dis. 2001;21:311–335. - PubMed
-
- Cassiman D, Barlow A, Vander Borght S, Libbrecht L, Pachnis V. Hepatic stellate cells do not derive from the neural crest. J Hepatol. 2006;44:1098–1104. - PubMed
-
- Hazra S, Xiong S, Wang J, Rippe RA, Krishna V, Chatterjee K, et al. Peroxisome proliferator-activated receptor γ induces a phenotypic switch from activated to quiescent hepatic stellate cells. J Biol Chem. 2004;279:11392–11401. - PubMed
-
- She H, Xiong S, Hazra S, Tsukamoto H. Adipogenic transcriptional regulation of hepatic stellate cells. J Biol Chem. 2005;280:4959–4967. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials
Miscellaneous
