Using VMD: an introductory tutorial
- PMID: 19085979
- PMCID: PMC2972669
- DOI: 10.1002/0471250953.bi0507s24
Using VMD: an introductory tutorial
Abstract
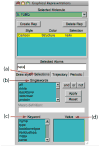
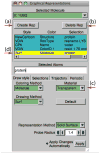
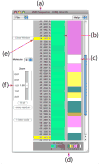
VMD (Visual Molecular Dynamics) is a molecular visualization and analysis program designed for biological systems such as proteins, nucleic acids, lipid bilayer assemblies, etc. This unit will serve as an introductory VMD tutorial. We will present several step-by-step examples of some of VMD's most popular features, including visualizing molecules in three dimensions with different drawing and coloring methods, rendering publication-quality figures, animating and analyzing the trajectory of a molecular dynamics simulation, scripting in the text-based Tcl/Tk interface, and analyzing both sequence and structure data for proteins.
Copyright 2008 by John Wiley & Sons, Inc.
Figures


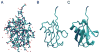

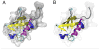










References
-
- Eastwood MP, Hardin C, Luthey-Schulten Z, Wolynes PG. Evaluating the protein structure-prediction schemes using energy landscape theory. IBM J Res Dev. 2001;45:475–497.
-
- Freddolino PL, Arkhipov AS, Larson SB, McPherson A, Schulten K. Molecular dynamics simulations of the complete satellite tobacco mosaic virus. Structure. 2006;14:437–449. - PubMed
-
- Frishman D, Argos P. Knowledge-based secondary structure assignment. Proteins. 1995;23:566–579. - PubMed
-
- Humphrey W, Dalke A, Schulten K. VMD – Visual Molecular Dynamics. J Mol Graphics. 1996;14:33–38. - PubMed
-
- Isralewitz B, Gao M, Schulten K. Steered molecular dynamics and mechanical functions of proteins. Curr Opin Struct Biol. 2001;11:224–230. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

