Environmental rRNA inventories miss over half of protistan diversity
- PMID: 19087295
- PMCID: PMC2625359
- DOI: 10.1186/1471-2180-8-222
Environmental rRNA inventories miss over half of protistan diversity
Abstract
Background: The main tool to discover novel microbial eukaryotes is the rRNA approach. This approach has important biases, including PCR discrimination against certain rRNA gene species, which makes molecular inventories skewed relative to the source communities. The degree of this bias has not been quantified, and it remains unclear whether species missed from clone libraries could be recovered by increasing sequencing efforts, or whether they cannot be detected in principle. Here we attempt to discriminate between these possibilities by statistically analysing four protistan inventories obtained using different general eukaryotic PCR primers.
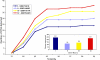
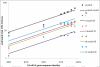
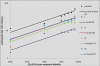
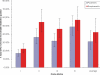
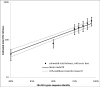
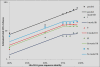
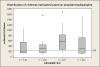
Results: We show that each PCR primer set-specific clone library is not a sample from the community diversity but rather from a fraction of this diversity. Therefore, even sequencing such clone libraries to saturation would only recover that fraction, which, according to the parametric models, varies between 17 +/- 4% to 49 +/- 10%, depending on the set of primers. The pooled data is thus qualitatively richer than individual libraries, even if normalized to the same sequencing effort.
Conclusion: The use of a single pair of primers leads to significant underestimation of the true community richness at all levels of taxonomic hierarchy. The majority of available protistan rRNA gene surveys likely sampled less than half of the target diversity, and might have completely missed the rest. The use of multiple PCR primers reduces this bias but does not necessarily eliminate it.
Figures

References
-
- Rusch DB, Halpern AL, Sutton G, Heidelberg KB, Williamson S, Yooseph S, Wu D, Eisen JA, Hoffman JM, Remington K, et al. The Sorcerer II Global Ocean Sampling expedition: northwest Atlantic through eastern tropical Pacific. PLoS Biol. 2007;5:e77. doi: 10.1371/journal.pbio.0050077. - DOI - PMC - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

