Upstream-binding factor is sequestered into herpes simplex virus type 1 replication compartments
- PMID: 19088274
- PMCID: PMC2885023
- DOI: 10.1099/vir.0.006353-0
Upstream-binding factor is sequestered into herpes simplex virus type 1 replication compartments
Abstract
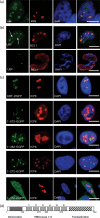
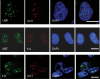
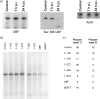
Previous reports have shown that adenovirus recruits nucleolar protein upstream-binding factor (UBF) into adenovirus DNA replication centres. Here, we report that despite having a different mode of viral DNA replication, herpes simplex virus type 1 (HSV-1) also recruits UBF into viral DNA replication centres. Moreover, as with adenovirus, enhanced green fluorescent protein-tagged fusion proteins of UBF inhibit viral DNA replication. We propose that UBF is recruited to the replication compartments to aid replication of HSV-1 DNA. In addition, this is a further example of the role of nucleolar components in viral life cycles.
Figures
Similar articles
-
Upstream binding factor inhibits herpes simplex virus replication.Virology. 2015 Sep;483:108-16. doi: 10.1016/j.virol.2015.04.003. Epub 2015 May 15. Virology. 2015. PMID: 25965800
-
Relocalization of upstream binding factor to viral replication compartments is UL24 independent and follows the onset of herpes simplex virus 1 DNA synthesis.J Virol. 2010 May;84(9):4810-5. doi: 10.1128/JVI.02437-09. Epub 2010 Feb 10. J Virol. 2010. PMID: 20147409 Free PMC article.
-
Nucleolar protein upstream binding factor is sequestered into adenovirus DNA replication centres during infection without affecting RNA polymerase I location or ablating rRNA synthesis.J Cell Sci. 2006 Jun 15;119(Pt 12):2621-31. doi: 10.1242/jcs.02982. J Cell Sci. 2006. PMID: 16763197
-
HSV-1 DNA Replication-Coordinated Regulation by Viral and Cellular Factors.Viruses. 2021 Oct 7;13(10):2015. doi: 10.3390/v13102015. Viruses. 2021. PMID: 34696446 Free PMC article. Review.
-
Molecular mechanisms of replication of herpes simplex virus 1.Acta Virol. 2000 Oct;44(5):289-307. Acta Virol. 2000. PMID: 11252674 Review.
Cited by
-
Nucleolar proteomics and viral infection.Proteomics. 2010 Nov;10(22):4077-86. doi: 10.1002/pmic.201000251. Proteomics. 2010. PMID: 20661956 Free PMC article. Review.
-
Involvement of the nucleolus in replication of human viruses.Rev Med Virol. 2009 Jul;19(4):201-14. doi: 10.1002/rmv.614. Rev Med Virol. 2009. PMID: 19399920 Free PMC article. Review.
-
Viruses and the nucleolus: the fatal attraction.Biochim Biophys Acta. 2014 Jun;1842(6):840-7. doi: 10.1016/j.bbadis.2013.12.010. Epub 2013 Dec 27. Biochim Biophys Acta. 2014. PMID: 24378568 Free PMC article.
-
HIV-1 infection causes a down-regulation of genes involved in ribosome biogenesis.PLoS One. 2014 Dec 2;9(12):e113908. doi: 10.1371/journal.pone.0113908. eCollection 2014. PLoS One. 2014. PMID: 25462981 Free PMC article.
-
Evidence for ubiquitin-regulated nuclear and subnuclear trafficking among Paramyxovirinae matrix proteins.PLoS Pathog. 2015 Mar 17;11(3):e1004739. doi: 10.1371/journal.ppat.1004739. eCollection 2015 Mar. PLoS Pathog. 2015. PMID: 25782006 Free PMC article.
References
-
- Bertrand, L. & Pearson, A. (2008). The conserved N-terminal domain of herpes simplex virus 1 UL24 protein is sufficient to induce the spatial redistribution of nucleolin. J Gen Virol 89, 1142–1151. - PubMed
-
- Besse, S. & Puvion-Dutilleul, F. (1996). Intranuclear retention of ribosomal RNAs in response to herpes simplex virus type 1 infection. J Cell Sci 109, 119–129. - PubMed
-
- Boehmer, P. E. & Lehman, I. R. (1997). Herpes simplex virus DNA replication. Annu Rev Biochem 66, 347–384. - PubMed
-
- Boisvert, F. M., van Koningsbruggen, S., Navascues, J. & Lamond, A. I. (2007). The multifunctional nucleolus. Nat Rev Mol Cell Biol 8, 574–585. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

