Cloning, Purification, and Partial Characterization of the Halobacterium sp. NRC-1 Minichromosome Maintenance (MCM) Helicase
- PMID: 19088906
- PMCID: PMC2593048
- DOI: 10.2174/1874285800802010013
Cloning, Purification, and Partial Characterization of the Halobacterium sp. NRC-1 Minichromosome Maintenance (MCM) Helicase
Abstract
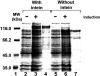
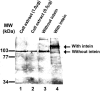
The MCM gene from the archaeon Halobacterium, with and without its intein, was cloned into an Escherichia coli expression vector, overexpressed and the protein was purified and antibodies were generated. The antibodies were used to demonstrate that in vivo only the processed enzyme, without the intein, could be detected.
Figures

References
-
- Grant WD. In: Halobacterium. 2 Grant GM, editor. Vol. 1. 2001.
-
- Kelman LM, Kelman Z. Archaea an archetype for replication initiation studies? Mol Microbiol. 2003;48:605–615. - PubMed
-
- Kelman Z, White MF. Archaeal DNA replication and repair. Curr Opin Microbiol. 2005;8:669–676. - PubMed
-
- Kasiviswanathan R, Shin JH, Melamud E, Kelman Z. Biochemical Characterization of the Methanothermobacter thermautotrophicus Minichromosome Maintenance (MCM) Helicase N-terminal Domains. J Biol Chem. 2004;279:28358–28366. - PubMed
LinkOut - more resources
Full Text Sources
