ERdj3, a luminal ER DnaJ homologue, binds directly to unfolded proteins in the mammalian ER: identification of critical residues
- PMID: 19090675
- PMCID: PMC2709649
- DOI: 10.1021/bi8015923
ERdj3, a luminal ER DnaJ homologue, binds directly to unfolded proteins in the mammalian ER: identification of critical residues
Abstract
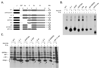
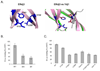
ERdj3 was identified as a soluble, lumenal DnaJ family member that binds to unassembled immunoglobulin heavy chains along with the BiP chaperone complex in the endoplasmic reticulum of mammalian cells. Here we demonstrated that ERdj3 binds directly to unfolded substrates. Secondary structure predictions suggested that the substrate binding domain of ERdj3 was likely to closely resemble Ydj1, a yeast cytosolic DnaJ family member, which was previously crystallized with a peptide bound to the C-terminal fragment composed of domains I, II, and III. Mutation of conserved residues in domain I, which formed the peptide binding site in Ydj1, affected ERdj3's substrate binding ability in mammalian cells and in vitro binding studies. Somewhat unexpectedly, we found that domain II, which is highly conserved among ERdj3 homologues, but very different from domain II of Ydj1, was also critical for substrate binding. In addition, we demonstrated that ERdj3 forms multimers in cells and found that the conserved carboxy-terminal residue phenylalanine 326 played a critical role in self-assembly. In vitro binding assays revealed that mutation of this residue to alanine diminished ERdj3's substrate binding ability, arguing that multimerization is important for substrate binding. Together, these studies demonstrate that the Ydj1 structure is conserved in another family member and reveal that among this group of DnaJ proteins domain II, which is not present in the closely related type II family members, also plays an essential role in substrate binding.
Figures



Similar articles
-
Dissection of structural and functional requirements that underlie the interaction of ERdj3 protein with substrates in the endoplasmic reticulum.J Biol Chem. 2014 Oct 3;289(40):27504-12. doi: 10.1074/jbc.M114.587147. Epub 2014 Aug 20. J Biol Chem. 2014. PMID: 25143379 Free PMC article.
-
The mammalian Hsp40 ERdj3 requires its Hsp70 interaction and substrate-binding properties to complement various yeast Hsp40-dependent functions.J Biol Chem. 2009 Nov 20;284(47):32462-71. doi: 10.1074/jbc.M109.000729. Epub 2009 Sep 11. J Biol Chem. 2009. PMID: 19748898 Free PMC article.
-
SDF2-like protein 1 (SDF2L1) regulates the endoplasmic reticulum localization and chaperone activity of ERdj3 protein.J Biol Chem. 2019 Dec 13;294(50):19335-19348. doi: 10.1074/jbc.RA119.009603. Epub 2019 Oct 17. J Biol Chem. 2019. PMID: 31624144 Free PMC article.
-
The diversity of the DnaJ/Hsp40 family, the crucial partners for Hsp70 chaperones.Cell Mol Life Sci. 2006 Nov;63(22):2560-70. doi: 10.1007/s00018-006-6192-6. Cell Mol Life Sci. 2006. PMID: 16952052 Free PMC article. Review.
-
Life and death of a BiP substrate.Semin Cell Dev Biol. 2010 Jul;21(5):472-8. doi: 10.1016/j.semcdb.2009.12.008. Epub 2009 Dec 21. Semin Cell Dev Biol. 2010. PMID: 20026282 Free PMC article. Review.
Cited by
-
Endoplasmic Reticulum Subproteome Analysis Reveals Underlying Defense Mechanisms of Wheat Seedling Leaves under Salt Stress.Int J Mol Sci. 2021 May 3;22(9):4840. doi: 10.3390/ijms22094840. Int J Mol Sci. 2021. PMID: 34063651 Free PMC article.
-
Dissection of structural and functional requirements that underlie the interaction of ERdj3 protein with substrates in the endoplasmic reticulum.J Biol Chem. 2014 Oct 3;289(40):27504-12. doi: 10.1074/jbc.M114.587147. Epub 2014 Aug 20. J Biol Chem. 2014. PMID: 25143379 Free PMC article.
-
Dual topology of co-chaperones at the membrane of the endoplasmic reticulum.Cell Death Discov. 2021 Aug 5;7(1):203. doi: 10.1038/s41420-021-00594-x. Cell Death Discov. 2021. PMID: 34354047 Free PMC article. Review.
-
Characterisation of Arabidopsis calnexin 1 and calnexin 2 in the endoplasmic reticulum and at plasmodesmata.Protoplasma. 2017 Jan;254(1):125-136. doi: 10.1007/s00709-015-0921-3. Epub 2015 Dec 17. Protoplasma. 2017. PMID: 26680228
-
Conformation transitions of the polypeptide-binding pocket support an active substrate release from Hsp70s.Nat Commun. 2017 Oct 31;8(1):1201. doi: 10.1038/s41467-017-01310-z. Nat Commun. 2017. PMID: 29084938 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

