VarDetect: a nucleotide sequence variation exploratory tool
- PMID: 19091032
- PMCID: PMC2638149
- DOI: 10.1186/1471-2105-9-S12-S9
VarDetect: a nucleotide sequence variation exploratory tool
Abstract
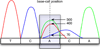
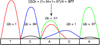
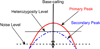
Background: Single nucleotide polymorphisms (SNPs) are the most commonly studied units of genetic variation. The discovery of such variation may help to identify causative gene mutations in monogenic diseases and SNPs associated with predisposing genes in complex diseases. Accurate detection of SNPs requires software that can correctly interpret chromatogram signals to nucleotides.
Results: We present VarDetect, a stand-alone nucleotide variation exploratory tool that automatically detects nucleotide variation from fluorescence based chromatogram traces. Accurate SNP base-calling is achieved using pre-calculated peak content ratios, and is enhanced by rules which account for common sequence reading artifacts. The proposed software tool is benchmarked against four other well-known SNP discovery software tools (PolyPhred, novoSNP, Genalys and Mutation Surveyor) using fluorescence based chromatograms from 15 human genes. These chromatograms were obtained from sequencing 16 two-pooled DNA samples; a total of 32 individual DNA samples. In this comparison of automatic SNP detection tools, VarDetect achieved the highest detection efficiency.
Availability: VarDetect is compatible with most major operating systems such as Microsoft Windows, Linux, and Mac OSX. The current version of VarDetect is freely available at http://www.biotec.or.th/GI/tools/vardetect.
Figures









References
-
- Uda M, Galanello R, Sanna S, Lettre G, Sankaran V, Chen W, Usala G, Busonero F, Maschio A, Albai G, et al. Genome-wide association study shows BCL11A associated with persistent fetal hemoglobin and amelioration of the phenotype of beta-thalassemia. Proc Natl Acad Sci USA. 2008;105:1620–1625. doi: 10.1073/pnas.0711566105. - DOI - PMC - PubMed
-
- Pandya G, Holmes M, Sunkara S, Sparks A, Bai Y, Verratti K, Saeed K, Venepally P, Jarrahi B, Fleischmann R, et al. A bioinformatic filter for improved base-call accuracy and polymorphism detection using the Affymetrix GeneChip whole-genome resequencing platform. Nucleic Acids Res. 2007;35:e148. doi: 10.1093/nar/gkm918. - DOI - PMC - PubMed
-
- Prosdocimi F, Lopes D, Peixoto F, Mourao M, Pacifico L, Ribeiro R, Ortega J. Effects of sample re-sequencing and trimming on the quality and size of assembled consensus sequences. Genet Mol Res. 2007;6:756–765. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

