Structural characterization of the Rous sarcoma virus RNA stability element
- PMID: 19091866
- PMCID: PMC2643715
- DOI: 10.1128/JVI.02113-08
Structural characterization of the Rous sarcoma virus RNA stability element
Abstract
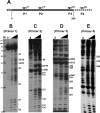
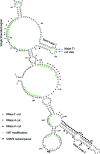
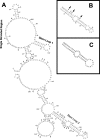
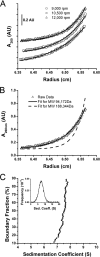
In eukaryotic cells, an mRNA bearing a premature termination codon (PTC) or an abnormally long 3' untranslated region (UTR) is often degraded by the nonsense-mediated mRNA decay (NMD) pathway. Despite the presence of a 5- to 7-kb 3' UTR, unspliced retroviral RNA escapes this degradation. We previously identified the Rous sarcoma virus (RSV) stability element (RSE), an RNA element downstream of the gag natural translation termination codon that prevents degradation of the unspliced viral RNA. Insertion of this element downstream of a PTC in the RSV gag gene also inhibits NMD. Using partial RNase digestion and selective 2'-hydroxyl acylation analyzed by primer extension (SHAPE) chemistry, we determined the secondary structure of this element. Incorporating RNase and SHAPE data into structural prediction programs definitively shows that the RSE contains an AU-rich stretch of about 30 single-stranded nucleotides near the 5' end and two substantial stem-loop structures. The overall secondary structure of the RSE appears to be conserved among 20 different avian retroviruses. The structural aspects of this element will serve as a tool in the future design of cis mutants in addressing the mechanism of stabilization.
Figures

References
-
- Amrani, N., R. Ganesan, S. Kervestin, D. A. Mangus, S. Ghosh, and A. Jacobson. 2004. A faux 3′-UTR promotes aberrant termination and triggers nonsense-mediated mRNA decay. Nature 432112-118. - PubMed
-
- Beemon, K. L. 2008. Retroviruses of birds, p. 455-459. In B. W. J. Mahy and M. H. V. Van Regenmortel (ed.), Encyclopedia of virology. Elsevier, Oxford, United Kingdom.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

