Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication
- PMID: 19091867
- PMCID: PMC2643704
- DOI: 10.1128/JVI.02371-08
Severe acute respiratory syndrome coronavirus protein 6 is required for optimal replication
Abstract
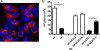
Severe acute respiratory syndrome coronavirus (SARS-CoV) encodes several accessory proteins of unknown function. One of these proteins, protein 6 (p6), which is encoded by ORF6, enhances virus replication when introduced into a heterologous murine coronavirus (mouse hepatitis virus [MHV]) but is not essential for optimal SARS-CoV replication after infection at a relatively high multiplicity of infection (MOI). Here, we reconcile these apparently conflicting results by showing that p6 enhances SARS-CoV replication to nearly the same extent as when expressed in the context of MHV if cells are infected at a low MOI and accelerates disease in mice transgenic for the human SARS-CoV receptor.
Figures

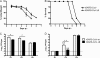

References
-
- Almazán, F., M. L. Dediego, C. Galan, D. Escors, E. Alvarez, J. Ortego, I. Sola, S. Zuniga, S. Alonso, J. L. Moreno, A. Nogales, C. Capiscol, and L. Enjuanes. 2006. Construction of a severe acute respiratory syndrome coronavirus infectious cDNA clone and a replicon to study coronavirus RNA synthesis. J. Virol. 8010900-10906. - PMC - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Research Materials
Miscellaneous

