The PDGF-C regulatory region SNP rs28999109 decreases promoter transcriptional activity and is associated with CL/P
- PMID: 19092777
- PMCID: PMC2788748
- DOI: 10.1038/ejhg.2008.245
The PDGF-C regulatory region SNP rs28999109 decreases promoter transcriptional activity and is associated with CL/P
Abstract
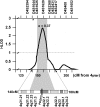
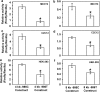
Human linkage and association studies suggest a gene(s) for nonsyndromic cleft lip with or without cleft palate (CL/P) on chromosome 4q31-q32 at or near the platelet-derived growth factor-C (PDGF-C) locus. The mouse Pdgfc(-/-) knockout shows that PDGF-C is essential for palatogenesis. To evaluate the role of PDGF-C in human clefting, we performed sequence analysis and SNP genotyping using 1048 multiplex CL/P families and 1000 case-control samples from multiple geographic origins. No coding region mutations were identified, but a novel -986 C>T SNP (rs28999109) was significantly associated with CL/P (P=0.01) in cases from Chinese families yielding evidence of linkage to 4q31-q32. Significant or near-significant association was also seen for this and several other PDGF-C SNPs in families from the United States, Spain, India, Turkey, China, and Colombia, whereas no association was seen in families from the Philippines, and Guatemala, and case-controls from Brazil. The -986T allele abolished six overlapping potential transcription regulatory motifs. Transfection assays of PDGF-C promoter reporter constructs show that the -986T allele is associated with a significant decrease (up to 80%) of PDGF-C gene promoter activity. This functional polymorphism acting on a susceptible genetic background may represent a component of human CL/P etiology.
Figures
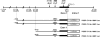
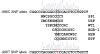

References
Publication types
MeSH terms
Substances
Grants and funding
- R37 DE008559/DE/NIDCR NIH HHS/United States
- N01 HG065403/HG/NHGRI NIH HHS/United States
- Z01 DE000711/ImNIH/Intramural NIH HHS/United States
- R01-DE012472/DE/NIDCR NIH HHS/United States
- R01 DE016148/DE/NIDCR NIH HHS/United States
- R01 DE012472/DE/NIDCR NIH HHS/United States
- R01-DE09886/DE/NIDCR NIH HHS/United States
- R00 DE018413/DE/NIDCR NIH HHS/United States
- R01 DE009886/DE/NIDCR NIH HHS/United States
- K99 DE018413/DE/NIDCR NIH HHS/United States
- R00 DE018085/DE/NIDCR NIH HHS/United States
- R01-DE016148/DE/NIDCR NIH HHS/United States
- R01 DE014667/DE/NIDCR NIH HHS/United States
- R37-DE08559/DE/NIDCR NIH HHS/United States
- R01-DE014667/DE/NIDCR NIH HHS/United States
- K02 DE015291/DE/NIDCR NIH HHS/United States
- P50 DE016215/DE/NIDCR NIH HHS/United States
- P50-DE016215/DE/NIDCR NIH HHS/United States
LinkOut - more resources
Full Text Sources
Medical
Molecular Biology Databases
Miscellaneous

