Combinatorial interaction between CCM pathway genes precipitates hemorrhagic stroke
- PMID: 19093037
- PMCID: PMC2590810
- DOI: 10.1242/dmm.000513
Combinatorial interaction between CCM pathway genes precipitates hemorrhagic stroke
Abstract
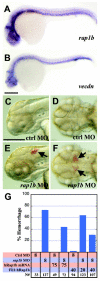
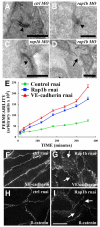
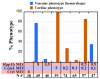
Intracranial hemorrhage (ICH) is a particularly severe form of stroke whose etiology remains poorly understood, with a highly variable appearance and onset of the disease (Felbor et al., 2006; Frizzell, 2005; Lucas et al., 2003). In humans, mutations in any one of three CCM genes causes an autosomal dominant genetic ICH disorder characterized by cerebral cavernous malformations (CCM). Recent evidence highlighting multiple interactions between the three CCM gene products and other proteins regulating endothelial junctional integrity suggests that minor deficits in these other proteins could potentially predispose to, or help to initiate, CCM, and that combinations of otherwise silent genetic deficits in both the CCM and interacting proteins might explain some of the variability in penetrance and expressivity of human ICH disorders. Here, we test this idea by combined knockdown of CCM pathway genes in zebrafish. Reducing the function of rap1b, which encodes a Ras GTPase effector protein for CCM1/Krit1, disrupts endothelial junctions in vivo and in vitro, showing it is a crucial player in the CCM pathway. Importantly, a minor reduction of Rap1b in combination with similar reductions in the products of other CCM pathway genes results in a high incidence of ICH. These findings support the idea that minor polygenic deficits in the CCM pathway can strongly synergize to initiate ICH.
Figures

References
-
- Cau J., Hall A. (2005). Cdc42 controls the polarity of the actin and microtubule cytoskeletons through two distinct signal transduction pathways. J. Cell Sci. 118, 2579–2587 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Molecular Biology Databases

