Heme-dependent metalloregulation by the iron response regulator (Irr) protein in Rhizobium and other Alpha-proteobacteria
- PMID: 19093075
- PMCID: PMC2659648
- DOI: 10.1007/s10534-008-9192-1
Heme-dependent metalloregulation by the iron response regulator (Irr) protein in Rhizobium and other Alpha-proteobacteria
Abstract
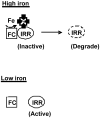
Perception and response to nutritional iron by bacteria is essential for viability, and for the ability to adapt to the environment. The iron response regulator (Irr) is part of a novel regulatory scheme employed by Rhizobium and other Alpha-Proteobacteria to control iron-dependent gene expression. Bradyrhizobium japonicum senses iron through the status of heme biosynthesis to regulate gene expression, thus it responds to an iron-dependent process rather than to iron directly. Irr mediates this response by interacting directly with ferrochelatase, the enzyme that catalyzes the final step in heme biosynthesis. Irr is expressed under iron limitation to both positively and negatively modulate gene expression, but degrades in response to direct binding to heme in iron-sufficient cells. Studies with Rhizobium reveal that the regulation of iron homeostasis in bacteria is more diverse than has been generally assumed.
Figures
Similar articles
-
Interaction between the bacterial iron response regulator and ferrochelatase mediates genetic control of heme biosynthesis.Mol Cell. 2002 Jan;9(1):155-62. doi: 10.1016/s1097-2765(01)00431-2. Mol Cell. 2002. PMID: 11804594
-
Living without Fur: the subtlety and complexity of iron-responsive gene regulation in the symbiotic bacterium Rhizobium and other alpha-proteobacteria.Biometals. 2007 Jun;20(3-4):501-11. doi: 10.1007/s10534-007-9085-8. Epub 2007 Feb 20. Biometals. 2007. PMID: 17310401 Review.
-
Heme binding to the second, lower-affinity site of the global iron regulator Irr from Rhizobium leguminosarum promotes oligomerization.FEBS J. 2011 Jun;278(12):2011-21. doi: 10.1111/j.1742-4658.2011.08117.x. Epub 2011 May 17. FEBS J. 2011. PMID: 21481185
-
Control of bacterial iron homeostasis by manganese.Proc Natl Acad Sci U S A. 2010 Jun 8;107(23):10691-5. doi: 10.1073/pnas.1002342107. Epub 2010 May 24. Proc Natl Acad Sci U S A. 2010. PMID: 20498065 Free PMC article.
-
Perception and Homeostatic Control of Iron in the Rhizobia and Related Bacteria.Annu Rev Microbiol. 2015;69:229-45. doi: 10.1146/annurev-micro-091014-104432. Epub 2015 Jul 16. Annu Rev Microbiol. 2015. PMID: 26195304 Review.
Cited by
-
Coordination chemistry of bacterial metal transport and sensing.Chem Rev. 2009 Oct;109(10):4644-81. doi: 10.1021/cr900077w. Chem Rev. 2009. PMID: 19788177 Free PMC article. Review. No abstract available.
-
Discovery of intracellular heme-binding protein HrtR, which controls heme efflux by the conserved HrtB-HrtA transporter in Lactococcus lactis.J Biol Chem. 2012 Feb 10;287(7):4752-8. doi: 10.1074/jbc.M111.297531. Epub 2011 Nov 14. J Biol Chem. 2012. PMID: 22084241 Free PMC article.
-
Signaling and Detoxification Strategies in Plant-Microbes Symbiosis under Heavy Metal Stress: A Mechanistic Understanding.Microorganisms. 2022 Dec 26;11(1):69. doi: 10.3390/microorganisms11010069. Microorganisms. 2022. PMID: 36677361 Free PMC article. Review.
-
Genomes from Uncultivated Pelagiphages Reveal Multiple Phylogenetic Clades Exhibiting Extensive Auxiliary Metabolic Genes and Cross-Family Multigene Transfers.mSystems. 2022 Oct 26;7(5):e0152221. doi: 10.1128/msystems.01522-21. Epub 2022 Aug 16. mSystems. 2022. PMID: 35972150 Free PMC article.
-
Differential control of Bradyrhizobium japonicum iron stimulon genes through variable affinity of the iron response regulator (Irr) for target gene promoters and selective loss of activator function.Mol Microbiol. 2014 May;92(3):609-24. doi: 10.1111/mmi.12584. Epub 2014 Apr 8. Mol Microbiol. 2014. PMID: 24646221 Free PMC article.
References
-
- Ahn BE, Cha J, Lee EJ, Han AR, Thompson CJ, Roe JH. Nur, a nickel-responsive regulator of the Fur family, regulates superoxide dismutases and nickel transport in Streptomyces coelicolor. Mol Microbiol. 2006;59:1848–1858. - PubMed
-
- Altuvia S, Almiron M, Huisman G, Kolter R, Storz G. The dps promoter is activated by OxyR during growth and by IHF and sigma S in stationary phase. Mol Microbiol. 1994;13:265–272. - PubMed
-
- Altuvia S, Weinstein-Fischer D, Zhang A, Postow L, Storz G. A small, stable RNA induced by oxidative stress: role as a pleiotropic regulator and antimutator. Cell. 1997;90:43–53. - PubMed
-
- Andrews SC, Robinson AK, Rodriguez-Quinones F. Bacterial iron homeostasis. FEMS Microbiol Rev. 2003;27:215–237. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources

