Elucidation of O-glycosylation structures of the beta-amyloid precursor protein by liquid chromatography-mass spectrometry using electron transfer dissociation and collision induced dissociation
- PMID: 19093876
- PMCID: PMC2743936
- DOI: 10.1021/pr800758g
Elucidation of O-glycosylation structures of the beta-amyloid precursor protein by liquid chromatography-mass spectrometry using electron transfer dissociation and collision induced dissociation
Erratum in
- J Proteome Res. 2009 Jul;8(7):3786. Alliquant, Bernadette [corrected to Allinquant, Bernadette]
Abstract
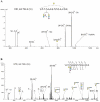
Accumulation and deposition of beta-amyloid peptide, a major constituent in neuritic plaques are hallmarks of Alzheimer's disease (AD) and AD-related neurodegenerative diseases. beta-Amyloid (Abeta) is derived from the proteolytic cleavage of amyloid precursor protein (APP), a transmembrane protein present in three major isoforms in brain comprising 695, 751 and 770 amino acids, respectively. Among other post-translational modifications, APP is modified during maturation by N- and O-glycosylation, which are thought to be responsible for its expression and secretion. Unlike N-glycosylation, no sites of O-glycosylation of APP have previously been reported. We report here the identification of three specific O-glycosylation sites of the secreted APP695 (sAPP695) produced in CHO cells, using a combination of high-performance liquid chromatography and electrospray-tandem mass spectrometry. With the use of electron transfer dissociation and collision induced dissociation (ETD and CID), we identified type, composition and structures of the Core 1 type O-linked glycans attached at the residues Thr 291, Thr 292 and Thr 576 of the full-length APP695. The glycosylations comprise multiple short glycans, containing N-acetyl galactosamine (GalNAc), Gal-GalNAc and sialic acid terminated structures. The presence of the glycopeptides in the tryptic mixture was identified using the CID-generated sugar oxonium ions. ETD proved to be valuable for the unambiguous identification of the modified sites as ETD fragmentation occurred along the peptide backbone with little or no cleavage of the glycans. Thus, the combination of the CID and ETD techniques in LC-MS is shown here, as a powerful tool for de novo identification of O-glycosylations at unknown modification sites in proteins.
Figures



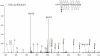




References
-
- Gralle M, Ferreira ST. Structure and functions of the human amyloid precursor protein: the whole is more than the sum of its parts. Prog Neurobiol. 2007;82(1):11–32. - PubMed
-
- Goldgaber D, Lerman MI, McBride OW, Saffiotti U, Gajdusek DC. Characterization and chromosomal localization of a cDNA encoding brain amyloid of Alzheimer’s disease. Science. 1987;235(4791):877–80. - PubMed
-
- Allinson TM, Parkin ET, Turner AJ, Hooper NM. ADAMs family members as amyloid precursor protein alpha-secretases. J Neurosci Res. 2003;74(3):342–52. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

