TREX2 exonuclease defective cells exhibit double-strand breaks and chromosomal fragments but not Robertsonian translocations
- PMID: 19094998
- PMCID: PMC2677549
- DOI: 10.1016/j.mrfmmm.2008.11.012
TREX2 exonuclease defective cells exhibit double-strand breaks and chromosomal fragments but not Robertsonian translocations
Abstract
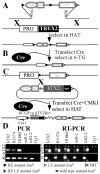
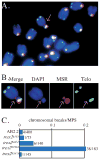
TREX2 is a 3'-->5' exonuclease that binds to DNA and removes 3' mismatched nucleotides. By an in vitro structure function analysis, we found a single amino acid change (H188A) completely ablates exonuclease activity and impairs DNA binding by about 60% while another change (R167A) impairs DNA binding by about 85% without impacting exonuclease activity. For a biological analysis, we generated trex2null cells by deleting the entire Trex2 coding sequences in mouse embryonic stem (ES) cells. We found Trex2 deletion caused high levels of Robertsonian translocations (RbTs) showing Trex2 is important for chromosomal maintenance. Here we evaluate the exonuclease and DNA binding domains by expressing in trex2(null) cells coding sequences for wild type human TREX2 (Trex2hTX2) or human TREX2 with the H188A change (Trex2H188A) or the R167A change (Trex2R167A). These cDNAs are positioned adjacent to the mouse Trex2 promoter by Cre-mediated knock-in. By observing metaphase spreads, we found Trex2H188A cells exhibited high levels of double-strand breaks (DSBs) and chromosomal fragments. Therefore, TREX2 may suppress spontaneous DSBs or exonuclease defective TREX2 may induce them in a dominate-negative manner. We also found Trex2hTX2, hTrex2H188A and hTrex2R167A cells did not exhibit RbTs. Thus, neither the exonuclease nor DNA binding domains suppress RbTs suggesting TREX2 possesses additional biochemical activities.
Figures
References
-
- Lindahl T, Gally JA, Edelman GM. Properties of deoxyribonuclease 3 from mammalian tissues. J Biol Chem. 1969;244:5014–5019. - PubMed
-
- Lindahl T. Excision of pyrimidine dimers from ultraviolet-irradiated DNA by exonucleases from mammalian cells. Eur J Biochem. 1971;18:407–414. - PubMed
-
- Mazur DJ, Perrino FW. Identification and expression of the TREX1 and TREX2 cDNA sequences encoding mammalian 3′-->5′ exonucleases. J Biol Chem. 1999;274:19655–19660. - PubMed
-
- Mazur DJ, Perrino FW. Excision of 3′ termini by the Trex1 and TREX2 3′-->5′ exonucleases. Characterization of the recombinant proteins. J Biol Chem. 2001;276:17022–17029. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

