SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs
- PMID: 19095803
- PMCID: PMC2632170
- DOI: 10.1101/gad.1717309
SMD and NMD are competitive pathways that contribute to myogenesis: effects on PAX3 and myogenin mRNAs
Abstract
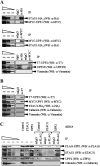
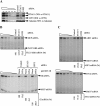
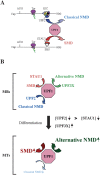
UPF1 functions in both Staufen 1 (STAU1)-mediated mRNA decay (SMD) and nonsense-mediated mRNA decay (NMD), which we show here are competitive pathways. STAU1- and UPF2-binding sites within UPF1 overlap so that STAU1 and UPF2 binding to UPF1 appear to be mutually exclusive. Furthermore, down-regulating the cellular abundance of STAU1, which inhibits SMD, increases the efficiency of NMD, whereas down-regulating the cellular abundance of UPF2, which inhibits NMD, increases the efficiency of SMD. Competition under physiological conditions is exemplified during the differentiation of C2C12 myoblasts to myotubes: The efficiency of SMD increases and the efficiency of NMD decreases, consistent with our finding that more STAU1 but less UPF2 bind UPF1 in myotubes compared with myoblasts. Moreover, an increase in the cellular level of UPF3X during myogenesis results in an increase in the efficiency of an alternative NMD pathway that, unlike classical NMD, is largely insensitive to UPF2 down-regulation. We discuss the remarkable balance between SMD and the two types of NMD in view of data indicating that PAX3 mRNA is an SMD target whose decay promotes myogenesis whereas myogenin mRNA is a classical NMD target encoding a protein required for myogenesis.
Figures




References
-
- Behm-Ansmant I., Kashima I., Rehwinkel J., Sauliere J., Wittkopp N., Izaurralde E. mRNA quality control: An ancient machinery recognizes and degrades mRNAs with nonsense codons. FEBS Lett. 2007;581:2845–2853. - PubMed
-
- Berger F.G., Szoka P. Biosynthesis of the major urinary proteins in mouse liver: A biochemical genetic study. Biochem. Genet. 1981;19:1261–1273. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
