A simple screen to identify promoters conferring high levels of phenotypic noise
- PMID: 19096504
- PMCID: PMC2588653
- DOI: 10.1371/journal.pgen.1000307
A simple screen to identify promoters conferring high levels of phenotypic noise
Abstract
Genetically identical populations of unicellular organisms often show marked variation in some phenotypic traits. To investigate the molecular causes and possible biological functions of this phenotypic noise, it would be useful to have a method to identify genes whose expression varies stochastically on a certain time scale. Here, we developed such a method and used it for identifying genes with high levels of phenotypic noise in Salmonella enterica ssp. I serovar Typhimurium (S. Typhimurium). We created a genomic plasmid library fused to a green fluorescent protein (GFP) reporter and subjected replicate populations harboring this library to fluctuating selection for GFP expression using fluorescent-activated cell sorting (FACS). After seven rounds of fluctuating selection, the populations were strongly enriched for promoters that showed a high amount of noise in gene expression. Our results indicate that the activity of some promoters of S. Typhimurium varies on such a short time scale that these promoters can absorb rapid fluctuations in the direction of selection, as imposed during our experiment. The genomic fragments that conferred the highest levels of phenotypic variation were promoters controlling the synthesis of flagella, which are associated with virulence and host-pathogen interactions. This confirms earlier reports that phenotypic noise may play a role in pathogenesis and indicates that these promoters have among the highest levels of noise in the S. Typhimurium genome. This approach can be applied to many other bacterial and eukaryotic systems as a simple method for identifying genes with noisy expression.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures
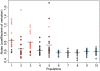
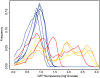
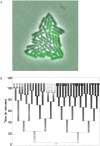
Similar articles
-
Bacterial genetics by flow cytometry: rapid isolation of Salmonella typhimurium acid-inducible promoters by differential fluorescence induction.Mol Microbiol. 1996 Oct;22(2):367-78. doi: 10.1046/j.1365-2958.1996.00120.x. Mol Microbiol. 1996. PMID: 8930920
-
Genetic Characterization of the Galactitol Utilization Pathway of Salmonella enterica Serovar Typhimurium.J Bacteriol. 2017 Jan 30;199(4):e00595-16. doi: 10.1128/JB.00595-16. Print 2017 Feb 15. J Bacteriol. 2017. PMID: 27956522 Free PMC article.
-
A Dopamine-Responsive Signal Transduction Controls Transcription of Salmonella enterica Serovar Typhimurium Virulence Genes.mBio. 2019 Apr 16;10(2):e02772-18. doi: 10.1128/mBio.02772-18. mBio. 2019. PMID: 30992361 Free PMC article.
-
The challenge of relating gene expression to the virulence of Salmonella enterica serovar Typhimurium.Curr Opin Biotechnol. 2011 Apr;22(2):200-10. doi: 10.1016/j.copbio.2011.02.007. Epub 2011 Mar 22. Curr Opin Biotechnol. 2011. PMID: 21388802 Review.
-
Non-genetic diversity shapes infectious capacity and host resistance.Trends Microbiol. 2012 Oct;20(10):461-6. doi: 10.1016/j.tim.2012.07.003. Epub 2012 Aug 10. Trends Microbiol. 2012. PMID: 22889945 Free PMC article. Review.
Cited by
-
The evolutionary emergence of stochastic phenotype switching in bacteria.Microb Cell Fact. 2011 Aug 30;10 Suppl 1(Suppl 1):S14. doi: 10.1186/1475-2859-10-S1-S14. Epub 2011 Aug 30. Microb Cell Fact. 2011. PMID: 21995592 Free PMC article.
-
A genome-wide analysis of promoter-mediated phenotypic noise in Escherichia coli.PLoS Genet. 2012 Jan;8(1):e1002443. doi: 10.1371/journal.pgen.1002443. Epub 2012 Jan 19. PLoS Genet. 2012. PMID: 22275871 Free PMC article.
-
The cost of virulence: retarded growth of Salmonella Typhimurium cells expressing type III secretion system 1.PLoS Pathog. 2011 Jul;7(7):e1002143. doi: 10.1371/journal.ppat.1002143. Epub 2011 Jul 28. PLoS Pathog. 2011. PMID: 21829349 Free PMC article.
-
Widespread inter-individual gene expression variability in Arabidopsis thaliana.Mol Syst Biol. 2019 Jan 24;15(1):e8591. doi: 10.15252/msb.20188591. Mol Syst Biol. 2019. PMID: 30679203 Free PMC article.
-
Pseudomonas syringae subpopulations cooperate by coordinating flagellar and type III secretion spatiotemporal dynamics to facilitate plant infection.Nat Microbiol. 2025 Apr;10(4):958-972. doi: 10.1038/s41564-025-01966-0. Epub 2025 Apr 2. Nat Microbiol. 2025. PMID: 40175722 Free PMC article.
References
-
- Ozbudak EM, Thattai M, Kurtser I, Grossman AD, van Oudenaarden A. Regulation of noise in the expression of a single gene. Nature Genetics. 2002;31:69–73. - PubMed
-
- Elowitz MB, Levine AJ, Siggia ED, Swain PS. Stochastic gene expression in a single cell. Science. 2002;297:1183–1186. - PubMed
-
- Acar M, Becskei A, van Oudenaarden A. Enhancement of cellular memory by reducing stochastic transitions. Nature. 2005;435:228–232. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources

