Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana
- PMID: 19096507
- PMCID: PMC2588661
- DOI: 10.1371/journal.pgen.1000285
Topoisomerase 3alpha and RMI1 suppress somatic crossovers and are essential for resolution of meiotic recombination intermediates in Arabidopsis thaliana
Abstract
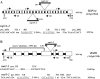
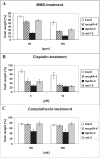
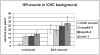
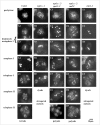
Topoisomerases are enzymes with crucial functions in DNA metabolism. They are ubiquitously present in prokaryotes and eukaryotes and modify the steady-state level of DNA supercoiling. Biochemical analyses indicate that Topoisomerase 3alpha (TOP3alpha) functions together with a RecQ DNA helicase and a third partner, RMI1/BLAP75, in the resolution step of homologous recombination in a process called Holliday Junction dissolution in eukaryotes. Apart from that, little is known about the role of TOP3alpha in higher eukaryotes, as knockout mutants show early lethality or strong developmental defects. Using a hypomorphic insertion mutant of Arabidopsis thaliana (top3alpha-2), which is viable but completely sterile, we were able to define three different functions of the protein in mitosis and meiosis. The top3alpha-2 line exhibits fragmented chromosomes during mitosis and sensitivity to camptothecin, suggesting an important role in chromosome segregation partly overlapping with that of type IB topoisomerases. Furthermore, AtTOP3alpha, together with AtRECQ4A and AtRMI1, is involved in the suppression of crossover recombination in somatic cells as well as DNA repair in both mammals and A. thaliana. Surprisingly, AtTOP3alpha is also essential for meiosis. The phenotype of chromosome fragmentation, bridges, and telophase I arrest can be suppressed by AtSPO11 and AtRAD51 mutations, indicating that the protein is required for the resolution of recombination intermediates. As Atrmi1 mutants have a similar meiotic phenotype to Attop3alpha mutants, both proteins seem to be involved in a mechanism safeguarding the entangling of homologous chromosomes during meiosis. The requirement of AtTOP3alpha and AtRMI1 in a late step of meiotic recombination strongly hints at the possibility that the dissolution of double Holliday Junctions via a hemicatenane intermediate is indeed an indispensable step of meiotic recombination.
Conflict of interest statement
The authors have declared that no competing interests exist.
Figures


References
Publication types
MeSH terms
Substances
Associated data
- Actions
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases
Research Materials

