Selective inhibition of carotenoid cleavage dioxygenases: phenotypic effects on shoot branching
- PMID: 19098002
- PMCID: PMC2643498
- DOI: 10.1074/jbc.M805453200
Selective inhibition of carotenoid cleavage dioxygenases: phenotypic effects on shoot branching
Abstract
Members of the carotenoid cleavage dioxygenase family catalyze the oxidative cleavage of carotenoids at various chain positions, leading to the formation of a wide range of apocarotenoid signaling molecules. To explore the functions of this diverse enzyme family, we have used a chemical genetic approach to design selective inhibitors for different classes of carotenoid cleavage dioxygenase. A set of 18 arylalkyl-hydroxamic acids was synthesized in which the distance between an iron-chelating hydroxamic acid and an aromatic ring was varied; these compounds were screened as inhibitors of four different enzyme classes, either in vitro or in vivo. Potent inhibitors were found that selectively inhibited enzymes that cleave carotenoids at the 9,10 position; 50% inhibition was achieved at submicromolar concentrations. Application of certain inhibitors at 100 microm to Arabidopsis node explants or whole plants led to increased shoot branching, consistent with inhibition of 9,10-cleavage.
Figures
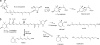
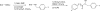
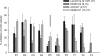

Similar articles
-
Biochemical characterization and selective inhibition of β-carotene cis-trans isomerase D27 and carotenoid cleavage dioxygenase CCD8 on the strigolactone biosynthetic pathway.FEBS J. 2015 Oct;282(20):3986-4000. doi: 10.1111/febs.13400. Epub 2015 Aug 31. FEBS J. 2015. PMID: 26257333
-
Characterization of three members of the Arabidopsis carotenoid cleavage dioxygenase family demonstrates the divergent roles of this multifunctional enzyme family.Plant J. 2006 Mar;45(6):982-93. doi: 10.1111/j.1365-313X.2006.02666.x. Plant J. 2006. PMID: 16507088
-
Enzymology of the carotenoid cleavage dioxygenases: reaction mechanisms, inhibition and biochemical roles.Arch Biochem Biophys. 2014 Feb 15;544:105-11. doi: 10.1016/j.abb.2013.10.005. Epub 2013 Oct 19. Arch Biochem Biophys. 2014. PMID: 24144525 Review.
-
MAX3/CCD7 is a carotenoid cleavage dioxygenase required for the synthesis of a novel plant signaling molecule.Curr Biol. 2004 Jul 27;14(14):1232-8. doi: 10.1016/j.cub.2004.06.061. Curr Biol. 2004. PMID: 15268852
-
Plant signaling: notes from the underground.Curr Biol. 2004 Nov 9;14(21):R929-30. doi: 10.1016/j.cub.2004.10.017. Curr Biol. 2004. PMID: 15530386 Review.
Cited by
-
Target sites for chemical regulation of strigolactone signaling.Front Plant Sci. 2014 Nov 5;5:623. doi: 10.3389/fpls.2014.00623. eCollection 2014. Front Plant Sci. 2014. PMID: 25414720 Free PMC article. Review.
-
Over-expression of the IGI1 leading to altered shoot-branching development related to MAX pathway in Arabidopsis.Plant Mol Biol. 2010 Aug;73(6):629-41. doi: 10.1007/s11103-010-9645-0. Epub 2010 May 15. Plant Mol Biol. 2010. PMID: 20473553 Free PMC article.
-
Isolation and functional characterization of two dioxygenases putatively involved in bixin biosynthesis in annatto (Bixa orellana L.).PeerJ. 2019 Jun 21;7:e7064. doi: 10.7717/peerj.7064. eCollection 2019. PeerJ. 2019. PMID: 31275744 Free PMC article.
-
Synthesis of Carlactone Derivatives to Develop a Novel Inhibitor of Strigolactone Biosynthesis.ACS Omega. 2023 Apr 5;8(15):13855-13862. doi: 10.1021/acsomega.3c00098. eCollection 2023 Apr 18. ACS Omega. 2023. PMID: 37091382 Free PMC article.
-
A cis-carotene derived apocarotenoid regulates etioplast and chloroplast development.Elife. 2020 Jan 31;9:e45310. doi: 10.7554/eLife.45310. Elife. 2020. PMID: 32003746 Free PMC article.
References
-
- Auldridge, M. E., McCarty, D. R., and Klee, H. J. (2006) Curr. Opin. Plant Biol. 9 315-321 - PubMed
-
- Zeevaart, J. A. D., and Creelman, R. A. (1988) Annu. Rev. Plant Physiol. Plant Mol. Biol. 39 439-473
-
- Schwartz, S. H., Tan, B. C., Gage, D. A., Zeevaart, J. A. D., and McCarty, D. R. (1997) Plant Physiol. 114 798-798
-
- von Lintig, J., and Vogt, K. (2000) J. Biol. Chem. 275 11915-11920 - PubMed
-
- Wyss, A., Wirtz, G., Woggon, W. D., Brugger, R., Wyss, M., Friedlein, A., Bachmann, H., and Hunziker, W. (2000) Biochem. Biophys. Res. Commun. 271 334-336 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

