Single-molecule DNA detection with an engineered MspA protein nanopore
- PMID: 19098105
- PMCID: PMC2634888
- DOI: 10.1073/pnas.0807514106
Single-molecule DNA detection with an engineered MspA protein nanopore
Abstract
Nanopores hold great promise as single-molecule analytical devices and biophysical model systems because the ionic current blockades they produce contain information about the identity, concentration, structure, and dynamics of target molecules. The porin MspA of Mycobacterium smegmatis has remarkable stability against environmental stresses and can be rationally modified based on its crystal structure. Further, MspA has a short and narrow channel constriction that is promising for DNA sequencing because it may enable improved characterization of short segments of a ssDNA molecule that is threaded through the pore. By eliminating the negative charge in the channel constriction, we designed and constructed an MspA mutant capable of electronically detecting and characterizing single molecules of ssDNA as they are electrophoretically driven through the pore. A second mutant with additional exchanges of negatively-charged residues for positively-charged residues in the vestibule region exhibited a factor of approximately 20 higher interaction rates, required only half as much voltage to observe interaction, and allowed ssDNA to reside in the vestibule approximately 100 times longer than the first mutant. Our results introduce MspA as a nanopore for nucleic acid analysis and highlight its potential as an engineerable platform for single-molecule detection and characterization applications.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

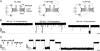
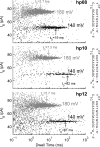

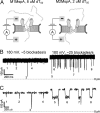
References
-
- Bezrukov SM, Vodyanoy I, Brutyan RA, Kasianowicz JJ. Dynamics and free energy of polymers partitioning into a nanoscale pore. Macromolecules. 1996;29:8517–8522.
-
- Braha O, et al. Designed protein pores as components for biosensors. Chem Biol. 1997;4:497–505. - PubMed
-
- Li J, et al. Ion-beam sculpting at nanometer-length scales. Nature. 2001;412:166–169. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

