TATA-binding protein variants that bypass the requirement for Mot1 in vivo
- PMID: 19098311
- PMCID: PMC2640957
- DOI: 10.1074/jbc.M808951200
TATA-binding protein variants that bypass the requirement for Mot1 in vivo
Abstract
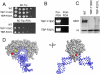
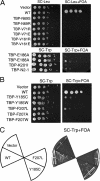
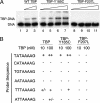
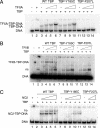
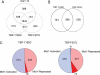
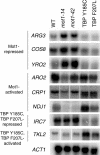
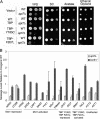
Mot1 is an essential TATA-binding protein (TBP)-associated factor and Snf2/Swi2 ATPase that both represses and activates transcription. Biochemical and structural results support a model in which ATP binding and hydrolysis induce a conformational change in Mot1 that drives local translocation along DNA, thus removing TBP. Although this activity explains transcriptional repression, it does not as easily explain Mot1-mediated transcriptional activation, and several different models have been proposed to explain how Mot1 activates transcription. To better understand the function of Mot1 in yeast cells in vivo, particularly with regard to gene activation, TBP mutants were identified that bypass the requirement for Mot1 in vivo. Although TBP has been extensively mutated and analyzed previously, this screen uncovered two novel TBP variants that are unique in their ability to bypass the requirement for Mot1. Surprisingly, in vitro analyses reveal that rather than having acquired an improved biochemical activity, one of the TBPs was defective for interaction with polymerase II preinitiation complex (PIC) components and other regulators of TBP function. The other mutant was defective for DNA binding in vitro yet was still recruited to chromatin in vivo. These results suggest that Mot1-mediated dissociation of TBP (or TBP-containing complexes) from chromatin can explain the Mot1 activation mechanism at some promoters. The results also suggest that PICs can be dynamically unstable and that appropriate PIC instability is critical for the regulation of transcription in vivo.
Figures
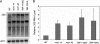
Similar articles
-
Function and structural organization of Mot1 bound to a natural target promoter.J Biol Chem. 2008 Sep 5;283(36):24935-48. doi: 10.1074/jbc.M803749200. Epub 2008 Jul 7. J Biol Chem. 2008. PMID: 18606810 Free PMC article.
-
Mot1-mediated control of transcription complex assembly and activity.EMBO J. 2005 May 4;24(9):1717-29. doi: 10.1038/sj.emboj.7600646. Epub 2005 Apr 7. EMBO J. 2005. PMID: 15861138 Free PMC article.
-
Molecular analysis of the SNF2/SWI2 protein family member MOT1, an ATP-driven enzyme that dissociates TATA-binding protein from DNA.Mol Cell Biol. 1997 Aug;17(8):4842-51. doi: 10.1128/MCB.17.8.4842. Mol Cell Biol. 1997. PMID: 9234740 Free PMC article.
-
One small step for Mot1; one giant leap for other Swi2/Snf2 enzymes?Biochim Biophys Acta. 2011 Sep;1809(9):488-96. doi: 10.1016/j.bbagrm.2011.05.012. Epub 2011 May 30. Biochim Biophys Acta. 2011. PMID: 21658482 Free PMC article. Review.
-
Roles for BTAF1 and Mot1p in dynamics of TATA-binding protein and regulation of RNA polymerase II transcription.Gene. 2003 Oct 2;315:1-13. doi: 10.1016/s0378-1119(03)00714-5. Gene. 2003. PMID: 14557059 Review.
Cited by
-
Independent RNA polymerase II preinitiation complex dynamics and nucleosome turnover at promoter sites in vivo.Genome Res. 2014 Jan;24(1):117-24. doi: 10.1101/gr.157792.113. Epub 2013 Dec 2. Genome Res. 2014. PMID: 24298073 Free PMC article.
-
How eukaryotic genes are transcribed.Crit Rev Biochem Mol Biol. 2009 Jun;44(2-3):117-41. doi: 10.1080/10409230902858785. Crit Rev Biochem Mol Biol. 2009. PMID: 19514890 Free PMC article. Review.
-
The basal initiation machinery: beyond the general transcription factors.Curr Opin Cell Biol. 2009 Jun;21(3):344-51. doi: 10.1016/j.ceb.2009.03.006. Epub 2009 May 4. Curr Opin Cell Biol. 2009. PMID: 19411170 Free PMC article. Review.
-
CTGC motifs within the HIV core promoter specify Tat-responsive pre-initiation complexes.Retrovirology. 2012 Jul 26;9:62. doi: 10.1186/1742-4690-9-62. Retrovirology. 2012. PMID: 22834489 Free PMC article.
-
RNA synthesis precision is regulated by preinitiation complex turnover.Genome Res. 2010 Dec;20(12):1679-88. doi: 10.1101/gr.109504.110. Epub 2010 Sep 20. Genome Res. 2010. PMID: 20855454 Free PMC article.
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

