Rng3, a member of the UCS family of myosin co-chaperones, associates with myosin heavy chains cotranslationally
- PMID: 19098712
- PMCID: PMC2637312
- DOI: 10.1038/embor.2008.228
Rng3, a member of the UCS family of myosin co-chaperones, associates with myosin heavy chains cotranslationally
Abstract
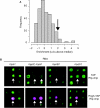
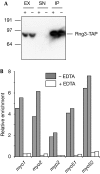
The production of functional myosin heavy chains in many eukaryotic organisms requires the function of proteins containing UCS domains (UNC-45/CRO1/She4), which bind to the myosin head domain and stimulate its folding. UCS proteins are essential for myosin-related functions such as muscle formation, RNA localization and cytokinesis. Here, we show that the Schizosaccharomyces pombe UCS protein Rng3 associates with polysomes, suggesting that UCS proteins might assist myosin folding cotranslationally. To identify Rng3 cotranslational targets systematically, we purified Rng3-associated RNAs and used DNA microarrays to identify the transcripts. Rng3 copurified with only seven transcripts (around 0.1% of S. pombe genes), including all five messenger RNAs encoding myosin heavy chains. These results suggest that every myosin heavy chain in S. pombe is a cotranslational target of Rng3. Furthermore, our data suggest that microarray-based approaches allow the genome-wide identification of cotranslational chaperone targets, and thus pave the way for the dissection of translation-linked chaperone networks.
Conflict of interest statement
The authors declare that they have no conflict of interest.
Figures

References
-
- Barral JM, Hutagalung AH, Brinker A, Hartl FU, Epstein HF (2002) Role of the myosin assembly protein UNC-45 as a molecular chaperone for myosin. Science 295: 669–671 - PubMed
-
- Bejsovec A, Anderson P (1988) Myosin heavy-chain mutations that disrupt Caenorhabditis elegans thick filament assembly. Genes Dev 2: 1307–1317 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

