An in vitro microfluidic approach to generating protein-interaction networks
- PMID: 19098921
- PMCID: PMC4117197
- DOI: 10.1038/nmeth.1289
An in vitro microfluidic approach to generating protein-interaction networks
Abstract
We developed an in vitro protein expression and interaction analysis platform based on a highly parallel and sensitive microfluidic affinity assay, and used it for 14,792 on-chip experiments, which exhaustively measured the protein-protein interactions of 43 Streptococcus pneumoniae proteins in quadruplicate. The resulting network of 157 interactions was denser than expected based on known networks. Analysis of the network revealed previously undescribed physical interactions among members of some biochemical pathways.
Figures
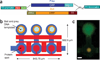


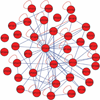
References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

