A genome-wide screen for modifiers of transgene variegation identifies genes with critical roles in development
- PMID: 19099580
- PMCID: PMC2646286
- DOI: 10.1186/gb-2008-9-12-r182
A genome-wide screen for modifiers of transgene variegation identifies genes with critical roles in development
Abstract
Background: Some years ago we established an N-ethyl-N-nitrosourea screen for modifiers of transgene variegation in the mouse and a preliminary description of the first six mutant lines, named MommeD1-D6, has been published. We have reported the underlying genes in three cases: MommeD1 is a mutation in SMC hinge domain containing 1 (Smchd1), a novel modifier of epigenetic gene silencing; MommeD2 is a mutation in DNA methyltransferase 1 (Dnmt1); and MommeD4 is a mutation in Smarca 5 (Snf2h), a known chromatin remodeler. The identification of Dnmt1 and Smarca5 attest to the effectiveness of the screen design.
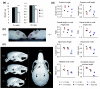
Results: We have now extended the screen and have identified four new modifiers, MommeD7-D10. Here we show that all ten MommeDs link to unique sites in the genome, that homozygosity for the mutations is associated with severe developmental abnormalities and that heterozygosity results in phenotypic abnormalities and reduced reproductive fitness in some cases. In addition, we have now identified the underlying genes for MommeD5 and MommeD10. MommeD5 is a mutation in Hdac1, which encodes histone deacetylase 1, and MommeD10 is a mutation in Baz1b (also known as Williams syndrome transcription factor), which encodes a transcription factor containing a PHD-type zinc finger and a bromodomain. We show that reduction in the level of Baz1b in the mouse results in craniofacial features reminiscent of Williams syndrome.
Conclusions: These results demonstrate the importance of dosage-dependent epigenetic reprogramming in the development of the embryo and the power of the screen to provide mouse models to study this process.
Figures






References
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

