Assessment of genetic and functional diversity of phosphate solubilizing fluorescent pseudomonads isolated from rhizospheric soil
- PMID: 19099598
- PMCID: PMC2625360
- DOI: 10.1186/1471-2180-8-230
Assessment of genetic and functional diversity of phosphate solubilizing fluorescent pseudomonads isolated from rhizospheric soil
Abstract
Background: Phosphorus is an essential macronutrient for the growth of plants. However, in most soils a large portion of phosphorus becomes insoluble and therefore, unavailable to plants. Knowledge on biodiversity of phosphate-solubilizing fluorescent pseudomonads is essential to understand their ecological role and their utilization in sustainable agriculture.
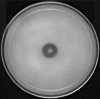
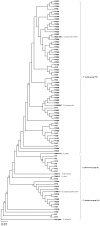
Results: Of 443 fluorescent pseudomonad strains tested, 80 strains (18%) showed positive for the solubilization of tri-calcium phosphate (Ca3(PO4)2) by the formation of visible dissolution halos on Pikovskaya's agar. These phosphate solubilizing strains showed high variability in utilizing various carbon sources. Numerical taxonomy of the phosphate solubilizing strains based on their carbon source utilization profiles resulted into three major phenons at a 0.76 similarity coefficient level. Genotypic analyses of strains by BOX (bacterial repetitive BOX element)-polymerase chain reaction (PCR) resulted into three distinct genomic clusters and 26 distinct BOX profiles at a 80% similarity level. On the basis of phenotypic characterization and 16S rRNA gene phylogenetic analyses strains were identified as Pseudomonas aeruginosa, P. mosselii, P. monteilii, P. plecoglossicida, P. putida, P. fulva and P. fluorescens. These phosphate solubilizing strains also showed the production of plant growth promoting enzymes, hormones and exhibited antagonism against phytopathogenic fungi that attack on various crops. Gene specific primers have identified the putative antibiotic producing strains. These putative strains were grown in fermentation media and production of antibiotics was confirmed by thin layer chromatography (TLC) and high performance liquid chromatography (HPLC).
Conclusion: Present study revealed a high degree of functional and genetic diversity among the phosphate solubilizing fluorescent pseudomonad bacteria. Due to their innate potential of producing an array of plant growth promoting enzymes, hormones and antifungal metabolites these phosphate solubilizing strains are considered to play a vital role in plant growth promotion, disease suppression and subsequent enhancement of yield.
Figures


References
-
- Richardson AE. Prospects for using soil microorganisms to improve the acquisition of phosphorus by plants. Aust J Plant Physiol. 2001;28:8797–906.
-
- Kim KY, Jordan D, McDonald GA. Effect of phosphate-solubilizing bacteria and vesicular-arbuscular mycorrhizeae on tomato growth and soil microbial activity. Biol Fertil Soils. 1998;26:79–87. doi: 10.1007/s003740050347. - DOI
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Miscellaneous

