The RepA_N replicons of Gram-positive bacteria: a family of broadly distributed but narrow host range plasmids
- PMID: 19100285
- PMCID: PMC2652615
- DOI: 10.1016/j.plasmid.2008.11.004
The RepA_N replicons of Gram-positive bacteria: a family of broadly distributed but narrow host range plasmids
Abstract
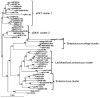
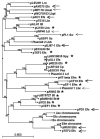
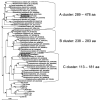
The pheromone-responsive conjugative plasmids of Enterococcus faecalis and the multiresistance plasmids pSK1 and pSK41 of Staphylococcus aureus are among the best studied plasmids native to Gram-positive bacteria. Although these plasmids seem largely restricted to their native hosts, protein sequence comparison of their replication initiator proteins indicates that they are clearly related. Homology searches indicate that these replicons are representatives of a large family of plasmids and a few phage that are widespread among the low G+C Gram-positive bacteria. We propose to name this family the RepA_N family of replicons after the annotated conserved domain that the initiator protein contains. Detailed sequence comparisons indicate that the initiator protein phylogeny is largely congruent with that of the host, suggesting that the replicons have evolved along with their current hosts and that intergeneric transfer has been rare. However, related proteins were identified on chromosomal regions bearing characteristics indicative of ICE elements, and the phylogeny of these proteins displayed evidence of more frequent intergeneric transfer. Comparison of stability determinants associated with the RepA_N replicons suggests that they have a modular evolution as has been observed in other plasmid families.
Figures



Similar articles
-
Replication of staphylococcal multiresistance plasmids.J Bacteriol. 2000 Apr;182(8):2170-8. doi: 10.1128/JB.182.8.2170-2178.2000. J Bacteriol. 2000. PMID: 10735859 Free PMC article.
-
Twenty years of the pPS10 replicon: insights on the molecular mechanism for the activation of DNA replication in iteron-containing bacterial plasmids.Plasmid. 2004 Sep;52(2):69-83. doi: 10.1016/j.plasmid.2004.06.002. Plasmid. 2004. PMID: 15336485 Review.
-
Analysis of the replication region of the cryptic plasmid pHE1 from the moderate halophile Halomonas elongata.Mol Gen Genet. 1999 Jun;261(4-5):851-61. doi: 10.1007/s004380050029. Mol Gen Genet. 1999. PMID: 10394923
-
Mosaic plasmids and mosaic replicons: evolutionary lessons from the analysis of genetic diversity in IncFII-related replicons.Microbiology (Reading). 2000 Sep;146 ( Pt 9):2267-2275. doi: 10.1099/00221287-146-9-2267. Microbiology (Reading). 2000. PMID: 10974114
-
[Replication control of iteron-containing plasmids--role of initiator protein-mediated DNA looping].Seikagaku. 1997 Nov;69(11):1272-7. Seikagaku. 1997. PMID: 9431017 Review. Japanese. No abstract available.
Cited by
-
Major families of multiresistant plasmids from geographically and epidemiologically diverse staphylococci.G3 (Bethesda). 2011 Dec;1(7):581-91. doi: 10.1534/g3.111.000760. Epub 2011 Dec 1. G3 (Bethesda). 2011. PMID: 22384369 Free PMC article.
-
Polyether ionophore resistance in a one health perspective.Front Microbiol. 2024 Jan 29;15:1347490. doi: 10.3389/fmicb.2024.1347490. eCollection 2024. Front Microbiol. 2024. PMID: 38351920 Free PMC article. Review.
-
A Glimpse into the World of Integrative and Mobilizable Elements in Streptococci Reveals an Unexpected Diversity and Novel Families of Mobilization Proteins.Front Microbiol. 2017 Mar 20;8:443. doi: 10.3389/fmicb.2017.00443. eCollection 2017. Front Microbiol. 2017. PMID: 28373865 Free PMC article.
-
Investigating the mobilome in clinically important lineages of Enterococcus faecium and Enterococcus faecalis.BMC Genomics. 2015 Apr 10;16:282. doi: 10.1186/s12864-015-1407-6. BMC Genomics. 2015. PMID: 25885771 Free PMC article.
-
The Plasmidomic Landscape of Clinical Methicillin-Resistant Staphylococcus aureus Isolates from Malaysia.Antibiotics (Basel). 2023 Apr 9;12(4):733. doi: 10.3390/antibiotics12040733. Antibiotics (Basel). 2023. PMID: 37107095 Free PMC article.
References
-
- Allignet J, El Solh N. Comparative analysis of staphylococcal plasmids carrying three streptogramin-resistance genes: vat-vgb-vga. Plasmid. 1999;42:134–138. - PubMed
-
- Aso Y, Koga H, Sashihara T, Nagao J-i, Kanemasa Y, Nakayama J, Sonomoto K. Description of complete DNA sequence of two plasmids from the nukacin ISK-1 producer, Staphylococcus warneri ISK-1. Plasmid. 2005;53:164–178. - PubMed
-
- Baba T, Takeuchi F, Kuroda M, Yuzawa H, Aoki K-i, Oguchi A, Nagai Y, Iwama N, Asano K, Naimi T, Kuroda H, Cui L, Yamamoto K, Hiramatsu K. Genome and virulence determinants of high virulence community-acquired MRSA. The Lancet. 2002;359:1819–1827. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

