Prediction of protein-protein binding site by using core interface residue and support vector machine
- PMID: 19102736
- PMCID: PMC2627892
- DOI: 10.1186/1471-2105-9-553
Prediction of protein-protein binding site by using core interface residue and support vector machine
Abstract
Background: The prediction of protein-protein binding site can provide structural annotation to the protein interaction data from proteomics studies. This is very important for the biological application of the protein interaction data that is increasing rapidly. Moreover, methods for predicting protein interaction sites can also provide crucial information for improving the speed and accuracy of protein docking methods.
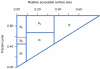
Results: In this work, we describe a binding site prediction method by designing a new residue neighbour profile and by selecting only the core-interface residues for SVM training. The residue neighbour profile includes both the sequential and the spatial neighbour residues of an interface residue, which is a more complete description of the physical and chemical characteristics surrounding the interface residue. The concept of core interface is applied in selecting the interface residues for training the SVM models, which is shown to result in better discrimination between the core interface and other residues. The best SVM model trained was tested on a test set of 50 randomly selected proteins. The sensitivity, specificity, and MCC for the prediction of the core interface residues were 60.6%, 53.4%, and 0.243, respectively. Our prediction results on this test set were compared with other three binding site prediction methods and found to perform better. Furthermore, our method was tested on the 101 unbound proteins from the protein-protein interaction benchmark v2.0. The sensitivity, specificity, and MCC of this test were 57.5%, 32.5%, and 0.168, respectively.
Conclusion: By improving both the descriptions of the interface residues and their surrounding environment and the training strategy, better SVM models were obtained and shown to outperform previous methods. Our tests on the unbound protein structures suggest further improvement is possible.
Figures
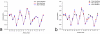
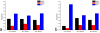
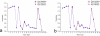

References
-
- Krogan NJ, Cagney G, Yu H, Zhong G, Guo X, Ignatchenko A, Li J, Pu S, Datta N, Tikuisis AP, Punna T, Peregrin-Alvarez JM, Shales M, Zhang X, Davery M, Robinson MD, Paccanaro A, Bray JE, Sheung A, Beattie B, Richards DP, Canadien V, Lalev A, Mena F, Wong P, Starostine A, Canete MM, Vlasblom J, Wu S, Orsi C, Collins SR, Chandran S, Haw R, Rilstone JJ, Gandi K, Thompson NJ, Musso SR, Onge PS, Ghanny S, Lam MH, Butland G, Altaf-UI AM, Kanaya S, Shilatifard A, O'Shea Weissman JS, Ingles CJ, Heghes TR, Parkinson J, Gerstein M, Wodak SJ, Emili A, Greenblatt JF. Global landscape of protein complexes in the yeast Saccharomyces cerevisiae. Nature. 2006;440:637–643. doi: 10.1038/nature04670. - DOI - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

