Complete genomic sequence analysis of infectious bronchitis virus Ark DPI strain and its evolution by recombination
- PMID: 19102764
- PMCID: PMC2628353
- DOI: 10.1186/1743-422X-5-157
Complete genomic sequence analysis of infectious bronchitis virus Ark DPI strain and its evolution by recombination
Abstract
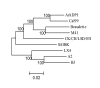
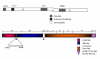
An infectious bronchitis virus Arkansas DPI (Ark DPI) virulent strain was sequenced, analyzed and compared with many different IBV strains and coronaviruses. The genome of Ark DPI consists of 27,620 nucleotides, excluding poly (A) tail, and comprises ten open reading frames. Comparative sequence analysis of Ark DPI with other IBV strains shows striking similarity to the Conn, Gray, JMK, and Ark 99, which were circulating during that time period. Furthermore, comparison of the Ark genome with other coronaviruses demonstrates a close relationship to turkey coronavirus. Among non-structural genes, the 5'untranslated region (UTR), 3C-like proteinase (3CLpro) and the polymerase (RdRp) sequences are 100% identical to the Gray strain. Among structural genes, S1 has 97% identity with Ark 99; S2 has 100% identity with JMK and 96% to Conn; 3b 99%, and 3C to N is 100% identical to Conn strain. Possible recombination sites were found at the intergenic region of spike gene, 3'end of S1 and 3a gene. Independent recombination events may have occurred in the entire genome of Ark DPI, involving four different IBV strains, suggesting that genomic RNA recombination may occur in any part of the genome at number of sites. Hence, we speculate that the Ark DPI strain originated from the Conn strain, but diverged and evolved independently by point mutations and recombination between field strains.
Figures

Similar articles
-
Emergence of subtype strains of the Arkansas serotype of infectious bronchitis virus in Delmarva broiler chickens.Avian Dis. 2000 Jul-Sep;44(3):568-81. Avian Dis. 2000. PMID: 11007004
-
Complete genome sequences of two avian infectious bronchitis viruses isolated in Egypt: Evidence for genetic drift and genetic recombination in the circulating viruses.Infect Genet Evol. 2017 Sep;53:7-14. doi: 10.1016/j.meegid.2017.05.006. Epub 2017 May 8. Infect Genet Evol. 2017. PMID: 28495648
-
Data from 11 years of molecular typing infectious bronchitis virus field isolates.Avian Dis. 2005 Dec;49(4):614-8. doi: 10.1637/7389-052905R.1. Avian Dis. 2005. PMID: 16405010
-
Identification of sequence changes responsible for the attenuation of avian infectious bronchitis virus strain Arkansas DPI.Arch Virol. 2009;154(3):495-9. doi: 10.1007/s00705-009-0325-9. Epub 2009 Feb 14. Arch Virol. 2009. PMID: 19219402 Free PMC article.
-
Molecular evolution and emergence of avian gammacoronaviruses.Infect Genet Evol. 2012 Aug;12(6):1305-11. doi: 10.1016/j.meegid.2012.05.003. Epub 2012 May 17. Infect Genet Evol. 2012. PMID: 22609285 Free PMC article. Review.
Cited by
-
Complete nucleotide analysis of the structural genome of the infectious bronchitis virus strain md27 reveals its mosaic nature.Viruses. 2009 Dec;1(3):1166-77. doi: 10.3390/v1031166. Epub 2009 Dec 4. Viruses. 2009. PMID: 21994587 Free PMC article.
-
Isolation of a novel serotype strain of infectious bronchitis virus ZZ2004 from ducks in China.Virus Genes. 2016 Oct;52(5):660-70. doi: 10.1007/s11262-016-1352-8. Epub 2016 May 10. Virus Genes. 2016. PMID: 27164844 Free PMC article.
-
S1 gene-based phylogeny of infectious bronchitis virus: An attempt to harmonize virus classification.Infect Genet Evol. 2016 Apr;39:349-364. doi: 10.1016/j.meegid.2016.02.015. Epub 2016 Feb 12. Infect Genet Evol. 2016. PMID: 26883378 Free PMC article.
-
A 25-Year-Old Sample Contributes the Complete Genome Sequence of Avian Coronavirus Vaccine Strain ArkDPI, Reisolated from Commercial Broilers in the United States.Microbiol Resour Announc. 2020 Feb 27;9(9):e00067-20. doi: 10.1128/MRA.00067-20. Microbiol Resour Announc. 2020. PMID: 32107295 Free PMC article.
-
Analysis of a QX-like avian infectious bronchitis virus genome identified recombination in the region containing the ORF 5a, ORF 5b, and nucleocapsid protein gene sequences.Virus Genes. 2013 Jun;46(3):454-64. doi: 10.1007/s11262-013-0884-4. Epub 2013 Jan 26. Virus Genes. 2013. PMID: 23355072 Free PMC article.
References
-
- Chomiak TW, Luginbuhl RE, Jungherr EL. The propagation and cytopathogenic effect of an egg-adapted strain of infectious bronchitis virus in tissue culture. Avian Dis. 1958;2:456–465. doi: 10.2307/1587486. - DOI
-
- Cavanagh D, Naqi S. Infectious Bronchitis. In: Saif YM, Barnes HJ, Glisson JR, Fadly AM, McDougald LR, Swayne DE, editor. Diseases of Poultry. 11. Ames: Iowa State University Press; 2003. pp. 101–120.
Publication types
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

