NUTS and BOLTS: applications of fluorescence-detected sedimentation
- PMID: 19103145
- PMCID: PMC2693311
- DOI: 10.1016/j.ab.2008.11.033
NUTS and BOLTS: applications of fluorescence-detected sedimentation
Abstract
Analytical ultracentrifugation is a widely used method for characterizing the solution behavior of macromolecules. However, the two commonly used detectors, absorbance and interference, impose some fundamental restrictions on the concentrations and complexity of the solutions that can be analyzed. The recent addition of a fluorescence detector for the XL-I analytical ultracentrifuge (AU-FDS) enables two different types of sedimentation experiments. First, the AU-FDS can detect picomolar concentrations of labeled solutes, allowing the characterization of very dilute solutions of macromolecules, applications we call normal use tracer sedimentation (NUTS). The great sensitivity of NUTS analysis allows the characterization of small quantities of materials and high-affinity interactions. Second, the AU-FDS allows characterization of trace quantities of labeled molecules in solutions containing high concentrations and complex mixtures of unlabeled molecules, applications we call biological on-line tracer sedimentation (BOLTS). The discrimination of BOLTS enables the size distribution of a labeled macromolecule to be determined in biological milieus such as cell lysates and serum. Examples that embody features of both NUTS and BOLTS applications are presented along with our observations on these applications.
Figures
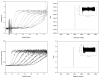

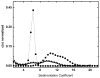
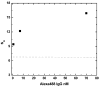
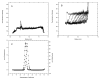
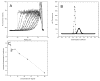



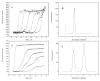
References
-
- Howlett GJ, Minton AP, Rivas G. Analytical ultracentrifugation for the study of protein association and assembly. Curr Opin Chem Biol. 2006;10:430–6. - PubMed
-
-
Laue, T.M. “Optical Systems of the XLA Ultracentrifuge” (1996) Applications Data Note for Spinco Division of Beckman Instruments, Inc. P.O.Box 10200, Palo Alto, California.
-
-
- MacGregor IK, Anderson AL, Laue TM. Fluorescence Detection for the XLI Ultracentrifuge. Biophys Chem. 2004;108:165–185. - PubMed
-
- Laue TM, Austin JB, Rau DA. A Light Intensity Measurement System for the Analytical Ultracentrifuge. Progr Colloid Polym Sci. 2006;131:1–8.
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

