CYP3A4 ubiquitination by gp78 (the tumor autocrine motility factor receptor, AMFR) and CHIP E3 ligases
- PMID: 19103148
- PMCID: PMC2881844
- DOI: 10.1016/j.abb.2008.12.001
CYP3A4 ubiquitination by gp78 (the tumor autocrine motility factor receptor, AMFR) and CHIP E3 ligases
Abstract
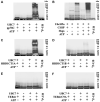
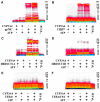
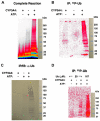
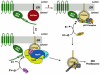
Human liver CYP3A4 is an endoplasmic reticulum (ER)-anchored hemoprotein responsible for the metabolism of >50% of clinically prescribed drugs. After heterologous expression in Saccharomyces cerevisiae, it is degraded via the ubiquitin (Ub)-dependent 26S proteasomal pathway that utilizes Ubc7p/Cue1p, but none of the canonical Ub-ligases (E3s) Hrd1p/Hrd3p, Doa10p, and Rsp5p involved in ER-associated degradation (ERAD). To identify an Ub-ligase capable of ubiquitinating CYP3A4, we examined various in vitro reconstituted mammalian E3 systems, using purified and functionally characterized recombinant components. Of these, the cytosolic domain of the ER-protein gp78, also known as the tumor autocrine motility factor receptor (AMFR), an UBC7-dependent polytopic RING-finger E3, effectively ubiquitinated CYP3A4 in vitro, as did the UbcH5a-dependent cytosolic E3 CHIP. CYP3A4 immunoprecipitation coupled with anti-Ub immunoblotting analyses confirmed its ubiquitination in these reconstituted systems. Thus, both UBC7/gp78 and UbcH5a/CHIP may be involved in CYP3A4 ERAD, although their relative physiological contribution remains to be established.
Figures

Similar articles
-
Hepatic cytochromes P450: structural degrons and barcodes, posttranslational modifications and cellular adapters in the ERAD-endgame.Drug Metab Rev. 2016 Aug;48(3):405-33. doi: 10.1080/03602532.2016.1195403. Epub 2016 Jun 20. Drug Metab Rev. 2016. PMID: 27320797 Free PMC article. Review.
-
Liver cytochrome P450 3A ubiquitination in vivo by gp78/autocrine motility factor receptor and C terminus of Hsp70-interacting protein (CHIP) E3 ubiquitin ligases: physiological and pharmacological relevance.J Biol Chem. 2010 Nov 12;285(46):35866-77. doi: 10.1074/jbc.M110.167189. Epub 2010 Sep 6. J Biol Chem. 2010. PMID: 20819951 Free PMC article.
-
Human liver cytochrome P450 3A4 ubiquitination: molecular recognition by UBC7-gp78 autocrine motility factor receptor and UbcH5a-CHIP-Hsc70-Hsp40 E2-E3 ubiquitin ligase complexes.J Biol Chem. 2015 Feb 6;290(6):3308-32. doi: 10.1074/jbc.M114.611525. Epub 2014 Dec 1. J Biol Chem. 2015. PMID: 25451919 Free PMC article.
-
Multisite phosphorylation of human liver cytochrome P450 3A4 enhances Its gp78- and CHIP-mediated ubiquitination: a pivotal role of its Ser-478 residue in the gp78-catalyzed reaction.Mol Cell Proteomics. 2012 Feb;11(2):M111.010132. doi: 10.1074/mcp.M111.010132. Epub 2011 Nov 17. Mol Cell Proteomics. 2012. PMID: 22101235 Free PMC article.
-
CHIP and gp78-mediated ubiquitination of CYP3A4: Implications for the pharmacology of anticancer agents.Cancer Biol Ther. 2011 Mar 15;11(6):549-51. doi: 10.4161/cbt.11.6.14834. Epub 2011 Mar 15. Cancer Biol Ther. 2011. PMID: 21270532 Free PMC article. Review.
Cited by
-
Membrane Protein Quantity Control at the Endoplasmic Reticulum.J Membr Biol. 2017 Aug;250(4):379-392. doi: 10.1007/s00232-016-9931-0. Epub 2016 Oct 14. J Membr Biol. 2017. PMID: 27743014 Free PMC article. Review.
-
Hepatic cytochromes P450: structural degrons and barcodes, posttranslational modifications and cellular adapters in the ERAD-endgame.Drug Metab Rev. 2016 Aug;48(3):405-33. doi: 10.1080/03602532.2016.1195403. Epub 2016 Jun 20. Drug Metab Rev. 2016. PMID: 27320797 Free PMC article. Review.
-
Cytochrome P450 endoplasmic reticulum-associated degradation (ERAD): therapeutic and pathophysiological implications.Acta Pharm Sin B. 2020 Jan;10(1):42-60. doi: 10.1016/j.apsb.2019.11.002. Epub 2019 Nov 8. Acta Pharm Sin B. 2020. PMID: 31993306 Free PMC article. Review.
-
Molecular basis for catalysis and substrate-mediated cellular stabilization of human tryptophan 2,3-dioxygenase.Sci Rep. 2016 Oct 20;6:35169. doi: 10.1038/srep35169. Sci Rep. 2016. PMID: 27762317 Free PMC article.
-
Ca2+/S100 proteins act as upstream regulators of the chaperone-associated ubiquitin ligase CHIP (C terminus of Hsc70-interacting protein).J Biol Chem. 2013 Mar 8;288(10):7158-68. doi: 10.1074/jbc.M112.436758. Epub 2013 Jan 23. J Biol Chem. 2013. PMID: 23344957 Free PMC article.
References
-
- Guengerich FP. In: Cytochrome P450: Structure, Mechanism and Biochemistry. Ortiz de Montellano P, editor. Kluwer, Academic/Plenum Press; New York: 2005. pp. 377–530.
-
- Correia MA. In: Cytochrome P450: Structure, Mechanism and Biochemistry. Ortiz de Montellano P, editor. Kluwer, Academic/Plenum Press; New York: 2005. pp. 619–657. and references therein.
-
- Watkins PB, Wrighton SA, Schuetz EG, Maurel P, Guzelian PS. J. Biol. Chem. 1986;261:6264–6271. - PubMed
-
- Eliasson E, Mkrtchian S, Halpert JR, Ingelman-Sundberg M. J. Biol. Chem. 1994;269:18378–18383. - PubMed
-
- Correia MA, Liao M. Expert Opin. Drug Metab. Toxicol. 2007;3:33–49. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

