Trypanosoma brucei spliced leader RNA maturation by the cap 1 2'-O-ribose methyltransferase and SLA1 H/ACA snoRNA pseudouridine synthase complex
- PMID: 19103757
- PMCID: PMC2643836
- DOI: 10.1128/MCB.01496-08
Trypanosoma brucei spliced leader RNA maturation by the cap 1 2'-O-ribose methyltransferase and SLA1 H/ACA snoRNA pseudouridine synthase complex
Abstract
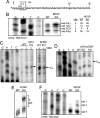
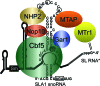
Kinetoplastid flagellates attach a 39-nucleotide spliced leader (SL) upstream of protein-coding regions in polycistronic RNA precursors through trans splicing. SL modifications include cap 2'-O-ribose methylation of the first four nucleotides and pseudouridine (psi) formation at uracil 28. In Trypanosoma brucei, TbMTr1 performs 2'-O-ribose methylation of the first transcribed nucleotide, or cap 1. We report the characterization of an SL RNA processing complex with TbMTr1 and the SLA1 H/ACA small nucleolar ribonucleoprotein (snoRNP) particle that guides SL psi(28) formation. TbMTr1 is in a high-molecular-weight complex containing the four conserved core proteins of H/ACA snoRNPs, a kinetoplastid-specific protein designated methyltransferase-associated protein (TbMTAP), and the SLA1 snoRNA. TbMTAP-null lines are viable but have decreased SL RNA processing efficiency in cap methylation, 3'-end maturation, and psi(28) formation. TbMTAP is required for association between TbMTr1 and the SLA1 snoRNP but does not affect U1 small nuclear RNA methylation. A complex methylation profile in the mRNA population of TbMTAP-null lines indicates an additional effect on cap 4 methylations. The TbMTr1 complex specializes the SLA1 H/ACA snoRNP for efficient processing of multiple modifications on the SL RNA substrate.
Figures




Similar articles
-
Hypermethylated cap 4 maximizes Trypanosoma brucei translation.Mol Microbiol. 2009 Jun;72(5):1100-10. doi: 10.1111/j.1365-2958.2009.06696.x. Mol Microbiol. 2009. PMID: 19504740 Free PMC article.
-
The 2'-O-ribose methyltransferase for cap 1 of spliced leader RNA and U1 small nuclear RNA in Trypanosoma brucei.Mol Cell Biol. 2007 Sep;27(17):6084-92. doi: 10.1128/MCB.00647-07. Epub 2007 Jul 2. Mol Cell Biol. 2007. PMID: 17606627 Free PMC article.
-
The TbMTr1 spliced leader RNA cap 1 2'-O-ribose methyltransferase from Trypanosoma brucei acts with substrate specificity.J Biol Chem. 2008 Feb 8;283(6):3161-3172. doi: 10.1074/jbc.M707367200. Epub 2007 Nov 29. J Biol Chem. 2008. PMID: 18048356
-
Unconventional rules of small nuclear RNA transcription and cap modification in trypanosomatids.Gene Expr. 2002;10(1-2):3-16. Gene Expr. 2002. PMID: 11868986 Free PMC article. Review.
-
mRNA splicing in trypanosomes.Int J Med Microbiol. 2012 Oct;302(4-5):221-4. doi: 10.1016/j.ijmm.2012.07.004. Epub 2012 Sep 7. Int J Med Microbiol. 2012. PMID: 22964417 Review.
Cited by
-
Genome-wide analysis of small nucleolar RNAs of Leishmania major reveals a rich repertoire of RNAs involved in modification and processing of rRNA.RNA Biol. 2015;12(11):1222-55. doi: 10.1080/15476286.2015.1038019. Epub 2015 May 13. RNA Biol. 2015. PMID: 25970223 Free PMC article.
-
Trypanosome cdc2-related kinase 9 controls spliced leader RNA cap4 methylation and phosphorylation of RNA polymerase II subunit RPB1.Mol Cell Biol. 2013 May;33(10):1965-75. doi: 10.1128/MCB.00156-13. Epub 2013 Mar 11. Mol Cell Biol. 2013. PMID: 23478263 Free PMC article.
-
The vault RNA of Trypanosoma brucei plays a role in the production of trans-spliced mRNA.J Biol Chem. 2019 Oct 25;294(43):15559-15574. doi: 10.1074/jbc.RA119.008580. Epub 2019 Aug 22. J Biol Chem. 2019. PMID: 31439669 Free PMC article.
-
Hypermethylated cap 4 maximizes Trypanosoma brucei translation.Mol Microbiol. 2009 Jun;72(5):1100-10. doi: 10.1111/j.1365-2958.2009.06696.x. Mol Microbiol. 2009. PMID: 19504740 Free PMC article.
-
Approaches for functional analysis of flagellar proteins in African trypanosomes.Methods Cell Biol. 2009;93:21-57. doi: 10.1016/S0091-679X(08)93002-8. Epub 2009 Dec 4. Methods Cell Biol. 2009. PMID: 20409810 Free PMC article.
References
-
- Arhin, G. K., E. Ullu, and C. Tschudi. 2006. 2′-O-Methylation of position 2 of the trypanosome spliced leader cap 4 is mediated by a 48 kDa protein related to vaccinia virus VP39. Mol. Biochem. Parasitol. 147137-139. - PubMed
-
- Bakin, A., and J. Ofengand. 1993. Four newly located pseudouridylate residues in Escherichia coli 23S ribosomal RNA are all at the peptidyl transferase center: analysis by the application of a new sequencing technique. Biochemistry 329754-9762. - PubMed
-
- Bangs, J. D., P. F. Crain, T. Hashizume, J. A. McCloskey, and J. C. Boothroyd. 1992. Mass spectrometry of mRNA cap 4 from trypanosomatids reveals two novel nucleosides. J. Biol. Chem. 2679805-9815. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials
Miscellaneous
