sarZ, a sarA family gene, is transcriptionally activated by MgrA and is involved in the regulation of genes encoding exoproteins in Staphylococcus aureus
- PMID: 19103928
- PMCID: PMC2648185
- DOI: 10.1128/JB.01555-08
sarZ, a sarA family gene, is transcriptionally activated by MgrA and is involved in the regulation of genes encoding exoproteins in Staphylococcus aureus
Abstract
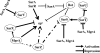
The expression of genes involved in the pathogenesis of Staphylococcus aureus is controlled by global regulatory loci, including two-component regulatory systems and transcriptional regulators (e.g., sar family genes). Most members of the SarA family have been partially characterized and shown to regulate a large numbers of target genes. Here, we describe the characterization of sarZ, a sarA paralog from S. aureus, and its regulatory relationship with other members of its family. Expression of sarZ was growth phase dependent with maximal expression in the early exponential phase of growth. Transcription of sarZ was reduced in an mgrA mutant and returned to a normal level in a complemented mgrA mutant strain, which suggests that mgrA acts as an activator of sarZ transcription. Purified MgrA protein bound to the sarZ promoter region, as determined by gel shift assays. Among the sarA family of genes analyzed, inactivation of sarZ increased sarS transcription, while it decreased agr transcription. The expression of potential target genes involved in virulence was evaluated in single and double mutants of sarZ with mgrA, sarX, and agr. Northern and zymogram analyses indicated that the sarZ gene product played a role in regulating several virulence genes, particularly those encoding exoproteins. Gel shift assays demonstrated nonspecific binding of purified SarZ protein to the promoter regions of the sarZ-regulated target genes. These results demonstrate the important role played by SarZ in controlling regulatory and virulence gene expression in S. aureus.
Figures





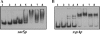
References
-
- Ausubel, F. M., R. Brent, R. E. Kingston, D. D. Moore, J. G. Seidman, J. A. Smith, and K. Struhl. 2000. Current protocols in molecular biology. John Wiley & Sons, New York, NY.
-
- Blevins, J. S., A. F. Gillaspy, T. M. Rechtin, B. K. Hurburt, and M. S. Smeltzer. 1999. The staphylococcal accessory regulator (sar) represses transcription of the S. aureus collagen adhesin gene (cna) in an agr-independent manner. Mol. Microbiol. 33317-326. - PubMed
-
- Brandenberger, M., M. Tschierske, P. Giachino, A. Wada, and B. Berger-Bachi. 2000. Inactivation of a novel three-cistronic operon tcaR-tcaA-tcaB increases teicoplanin resistance in Staphylococcus aureus. Biochim. Biophy. Acta 1523135-139. - PubMed
-
- Cassat, J., P. M. Dunman, E. Murphy, S. J. Projan, K. E. Beenken, K. J. Palm, S.-J. Yang, K. C. Rice, K. W. Bayles, and M. S. Smeltzer. 2006. Transcriptional profiling of a Staphylococcus aureus clinical isolate and its isogenic agr and sarA mutants reveals global differences in comparison to the laboratory strain RN6390. Microbiology 1523075-3090. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Miscellaneous

