A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes
- PMID: 19104045
- PMCID: PMC2606902
- DOI: 10.1073/pnas.0810772105
A large-scale analysis of tissue-specific pathology and gene expression of human disease genes and complexes
Abstract
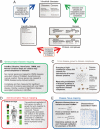
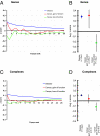
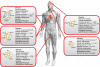
Heritable diseases are caused by germ-line mutations that, despite tissuewide presence, often lead to tissue-specific pathology. Here, we make a systematic analysis of the link between tissue-specific gene expression and pathological manifestations in many human diseases and cancers. Diseases were systematically mapped to tissues they affect from disease-relevant literature in PubMed to create a disease-tissue covariation matrix of high-confidence associations of >1,000 diseases to 73 tissues. By retrieving >2,000 known disease genes, and generating 1,500 disease-associated protein complexes, we analyzed the differential expression of a gene or complex involved in a particular disease in the tissues affected by the disease, compared with nonaffected tissues. When this analysis is scaled to all diseases in our dataset, there is a significant tendency for disease genes and complexes to be overexpressed in the normal tissues where defects cause pathology. In contrast, cancer genes and complexes were not overexpressed in the tissues from which the tumors emanate. We specifically identified a complex involved in XY sex reversal that is testis-specific and down-regulated in ovaries. We also identified complexes in Parkinson disease, cardiomyopathies, and muscular dystrophy syndromes that are similarly tissue specific. Our method represents a conceptual scaffold for organism-spanning analyses and reveals an extensive list of tissue-specific draft molecular pathways, both known and unexpected, that might be disrupted in disease.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

References
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical

