Properties of developmental gene regulatory networks
- PMID: 19104053
- PMCID: PMC2629280
- DOI: 10.1073/pnas.0806007105
Properties of developmental gene regulatory networks
Abstract
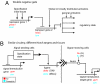
The modular components, or subcircuits, of developmental gene regulatory networks (GRNs) execute specific developmental functions, such as the specification of cell identity. We survey examples of such subcircuits and relate their structures to corresponding developmental functions. These relations transcend organisms and genes, as illustrated by the similar structures of the subcircuits controlling the specification of the mesectoderm in the Drosophila embryo and the endomesoderm in the sea urchin, even though the respective subcircuits are composed of nonorthologous regulatory genes.
Conflict of interest statement
The authors declare no conflict of interest.
Figures
References
-
- Davidson EH. The Regulatory Genome. Gene Regulatory Networks in Development and Evolution. San Diego: Academic; 2006.
-
- De Renzis S, Yu J, Zinzen R, Wieschaus E. Dorsal-ventral pattern of Delta trafficking is established by a Snail–Tom–Neuralized pathway. Dev Cell. 2006;10:257–264. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

