A gene regulatory network armature for T lymphocyte specification
- PMID: 19104054
- PMCID: PMC2629331
- DOI: 10.1073/pnas.0806501105
A gene regulatory network armature for T lymphocyte specification
Abstract
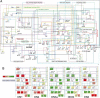
Choice of a T lymphoid fate by hematopoietic progenitor cells depends on sustained Notch-Delta signaling combined with tightly regulated activities of multiple transcription factors. To dissect the regulatory network connections that mediate this process, we have used high-resolution analysis of regulatory gene expression trajectories from the beginning to the end of specification, tests of the short-term Notch dependence of these gene expression changes, and analyses of the effects of overexpression of two essential transcription factors, namely PU.1 and GATA-3. Quantitative expression measurements of >50 transcription factor and marker genes have been used to derive the principal components of regulatory change through which T cell precursors progress from primitive multipotency to T lineage commitment. Our analyses reveal separate contributions of Notch signaling, GATA-3 activity, and down-regulation of PU.1. Using BioTapestry (www.BioTapestry.org), the results have been assembled into a draft gene regulatory network for the specification of T cell precursors and the choice of T as opposed to myeloid/dendritic or mast-cell fates. This network also accommodates effects of E proteins and mutual repression circuits of Gfi1 against Egr-2 and of TCF-1 against PU.1 as proposed elsewhere, but requires additional functions that remain unidentified. Distinctive features of this network structure include the intense dose dependence of GATA-3 effects, the gene-specific modulation of PU.1 activity based on Notch activity, the lack of direct opposition between PU.1 and GATA-3, and the need for a distinct, late-acting repressive function or functions to extinguish stem and progenitor-derived regulatory gene expression.
Conflict of interest statement
The authors declare no conflict of interest.
Figures

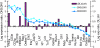
References
-
- Oliveri P, Davidson EH. Development: Built to run, not fail. Science. 2007;315:1510–1511. - PubMed
-
- Laslo P, et al. Multilineage transcriptional priming and determination of alternate hematopoietic cell fates. Cell. 2006;126:755–766. - PubMed
-
- Rothenberg EV. Negotiation of the T lineage fate decision by transcription-factor interplay and microenvironmental signals. Immunity. 2007;26:690–702. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Molecular Biology Databases

