Genomic organization and expression demonstrate spatial and temporal Hox gene colinearity in the lophotrochozoan Capitella sp. I
- PMID: 19104667
- PMCID: PMC2603591
- DOI: 10.1371/journal.pone.0004004
Genomic organization and expression demonstrate spatial and temporal Hox gene colinearity in the lophotrochozoan Capitella sp. I
Abstract
Hox genes define regional identities along the anterior-posterior axis in many animals. In a number of species, Hox genes are clustered in the genome, and the relative order of genes corresponds with position of expression in the body. Previous Hox gene studies in lophotrochozoans have reported expression for only a subset of the Hox gene complement and/or lack detailed genomic organization information, limiting interpretations of spatial and temporal colinearity in this diverse animal clade. We studied expression and genomic organization of the single Hox gene complement in the segmented polychaete annelid Capitella sp. I. Total genome searches identified 11 Hox genes in Capitella, representing 11 distinct paralog groups thought to represent the ancestral lophotrochozoan complement. At least 8 of the 11 Capitella Hox genes are genomically linked in a single cluster, have the same transcriptional orientation, and lack interspersed non-Hox genes. Studying their expression by situ hybridization, we find that the 11 Capitella Hox genes generally exhibit spatial and temporal colinearity. With the exception of CapI-Post1, Capitella Hox genes are all expressed in broad ectodermal domains during larval development, consistent with providing positional information along the anterior-posterior axis. The anterior genes CapI-lab, CapI-pb, and CapI-Hox3 initiate expression prior to the appearance of segments, while more posterior genes appear at or soon after segments appear. Many of the Capitella Hox genes have either an anterior or posterior expression boundary coinciding with the thoracic-abdomen transition, a major body tagma boundary. Following metamorphosis, several expression patterns change, including appearance of distinct posterior boundaries and restriction to the central nervous system. Capitella Hox genes have maintained a clustered organization, are expressed in the canonical anterior-posterior order found in other metazoans, and exhibit spatial and temporal colinearity, reflecting Hox gene characteristics that likely existed in the protostome-deuterostome ancestor.
Conflict of interest statement
Figures














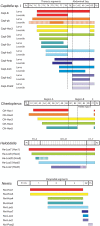
References
-
- Carroll SB. Homeotic genes and the evolution of arthropods and chordates. Nature. 1995;376:479–485. - PubMed
-
- Hughes CL, Kaufman TC. Hox genes and the evolution of the arthropod body plan. Evol Dev. 2002;4:459–499. - PubMed
-
- Duboule D. The rise and fall of Hox gene clusters. Development. 2007;134:2549–2560. - PubMed
-
- Krumlauf R. Hox genes in vertebrate development. Cell. 1994;78:191–201. - PubMed
-
- Ferrier DE, Holland PW. Ancient origin of the Hox gene cluster. Nat Rev Genet. 2001;2:33–38. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous

