Complete genome sequence of Nariva virus, a rodent paramyxovirus
- PMID: 19104752
- PMCID: PMC7086651
- DOI: 10.1007/s00705-008-0287-3
Complete genome sequence of Nariva virus, a rodent paramyxovirus
Abstract
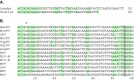
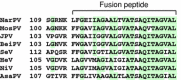
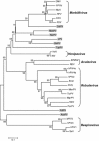
Nariva virus (NarPV) was isolated from forest rodents (Zygodontomys b. brevicauda) in eastern Trinidad in the early 1960s. Initial classification within the family Paramyxoviridae was based mainly on morphological observations including the structure of nucleocapsids and virion surface projections. Here, we report the characterization of the complete genome sequence of NarPV. The genome is 15,276 nucleotides in length, conforming to the rule-of-six, and has a genome organization typical of most members of the family, with six transcriptional units in the order 3'-N-P-M-F-H-L-5'. The gene junctions contain highly conserved gene start and stop signals and a tri-nucleotide intergenic sequence present in most members of the subfamily Paramyxovirinae. Sequence comparison studies indicate that NarPV is most closely related to Mossman virus, which was isolated from wild rats (Rattus leucopus) in Queensland, Australia, in 1970. This study confirmed the classification of NarPV as a member of the subfamily Paramyxovirinae and established the close genome organization and sequence relationship between the two rodent paramyxoviruses isolated almost a decade apart and from two locations separated by more than 15,000 km.
Figures


References
-
- Adkins RM, Gelke EL, Rowe D, Honeycutt RL. Molecular phylogeny and divergence time estimates for major rodent groups: evidence from multiple genes. Mol Biol Evol. 2001;18:777–791. - PubMed
MeSH terms
Substances
Associated data
- Actions
LinkOut - more resources
Full Text Sources

