A comprehensive analysis of Trypanosoma brucei mitochondrial proteome
- PMID: 19105172
- PMCID: PMC2869593
- DOI: 10.1002/pmic.200800477
A comprehensive analysis of Trypanosoma brucei mitochondrial proteome
Abstract
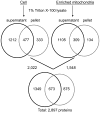
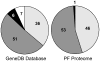
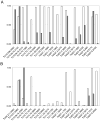
The composition of the large, single, mitochondrion (mt) of Trypanosoma brucei was characterized by MS (2-D LC-MS/MS and gel-LC-MS/MS) analyses. A total of 2897 proteins representing a substantial proportion of procyclic form cellular proteome were identified, which confirmed the validity of the vast majority of gene predictions. The data also showed that the genes annotated as hypothetical (species specific) were overpredicted and that virtually all genes annotated as hypothetical, unlikely are not expressed. By comparing the MS data with genome sequence, 40 genes were identified that were not previously predicted. The data are placed in a publicly available web-based database (www.TrypsProteome.org). The total mitochondrial proteome is estimated at 1008 proteins, with 401, 196, and 283 assigned to the mt with high, moderate, and lower confidence, respectively. The remaining mitochondrial proteins were estimated by statistical methods although individual assignments could not be made. The identified proteins have predicted roles in macromolecular, metabolic, energy generating, and transport processes providing a comprehensive profile of the protein content and function of the T. brucei mt.
Figures
References
-
- Berriman M, Ghedin E, Hertz-Fowler C, Blandin G, et al. The genome of the African trypanosome, Trypanosoma brucei. Science. 2005;309:416–422. - PubMed
-
- El-Sayed NMA, Myler PJ, Bartholomeu D, Nilsson D, et al. The genome sequence of Trypanosoma cruzi, etiological agent of Chagas’ disease. Science. 2005;309(5733):409–415. - PubMed
-
- El-Sayed NMA, Myler PJ, Blandin G, Berriman M, et al. Comparative genomics of trypanosomatid parasitic protozoa. Science. 2005;309(5733):404–409. - PubMed
-
- Martinez-Calvillo S, Yan S, Nguyen D, Fox M, et al. Transcription of Leishmania major Friedlin chromosome 1 initiates in both directions within a single region. Mol Cell. 2003;11(5):1291–1299. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases

