Mass spectrometry based targeted protein quantification: methods and applications
- PMID: 19105742
- PMCID: PMC2657955
- DOI: 10.1021/pr800538n
Mass spectrometry based targeted protein quantification: methods and applications
Abstract
The recent advance in technology for mass spectrometry-based targeted protein quantification has opened new avenues for a broad range of proteomic applications in clinical research. The major breakthroughs are highlighted by the capability of using a "universal" approach to perform quantitative assays for a wide spectrum of proteins with minimum restrictions and the ease of assembling multiplex detections in a single measurement. The quantitative approach relies on the use of synthetic stable isotope labeled peptides or proteins, which precisely mimic their endogenous counterparts and act as internal standards to quantify the corresponding candidate proteins. This report reviews recently developed platform technologies for emerging applications of clinical proteomics and biomarker development.
Figures
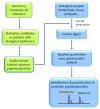

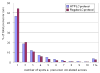
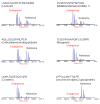
References
-
- Aebersold R, Mann M. Mass spectrometry-based proteomics. Nature. 2003;422(6928):198–207. - PubMed
-
- Ali M, Manolios N. Proteomics in rheumatology: a new direction for old diseases. Semin Arthritis Rheum. 2005;35(2):67–76. - PubMed
-
- Alpantaki K, Tsiridis E, Pape HC, Giannoudis PV. Application of clinical proteomics in diagnosis and management of trauma patients. Injury. 2007;38(3):263–271. - PubMed
-
- Ardekani AM, Liotta LA, Petricoin EF., III Clinical potential of proteomics in the diagnosis of ovarian cancer. Expert Rev Mol Diagn. 2002;2(4):312–320. - PubMed
-
- Bertucci F, Birnbaum D, Goncalves A. Proteomics of breast cancer: principles and potential clinical applications. Mol Cell Proteomics. 2006;5(10):1772–1786. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

