Comparative secretome analyses of two Trichoderma reesei RUT-C30 and CL847 hypersecretory strains
- PMID: 19105830
- PMCID: PMC2631499
- DOI: 10.1186/1754-6834-1-18
Comparative secretome analyses of two Trichoderma reesei RUT-C30 and CL847 hypersecretory strains
Abstract
Background: Due to its capacity to produce large amounts of cellulases, Trichoderma reesei is increasingly been researched in various fields of white biotechnology, especially in biofuel production from lignocellulosic biomass. The commercial enzyme mixtures produced at industrial scales are not well characterized, and their proteinaceous components are poorly identified and quantified. The development of proteomic methods has made it possible to comprehensively overview the enzymes involved in lignocellulosic biomass degradation which are secreted under various environmental conditions.
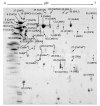
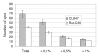
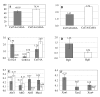
Results: The protein composition of the secretome produced by industrial T. reesei (strain CL847) grown on a medium promoting the production of both cellulases and hemicellulases was explored using two-dimensional electrophoresis and MALDI-TOF or LC-MS/MS protein identification. A total of 22 protein species were identified. As expected, most of them are potentially involved in biomass degradation. The 2D map obtained was then used to compare the secretomes produced by CL847 and another efficient cellulolytic T. reesei strain, Rut-C30, the reference cellulase-overproducing strain using lactose as carbon source and inducer of cellulases.
Conclusion: This study provides the most complete mapping of the proteins secreted by T. reesei to date. We report on the first use of proteomics to compare secretome composition between two cellulase-overproducing strains Rut-C30 and CL847 grown under similar conditions. Comparison of protein patterns in both strains highlighted many unexpected differences between cellulase cocktails. The results demonstrate that 2D electrophoresis is a promising tool for studying cellulase production profiles, whether for industrial characterization of an entire secretome or for a more fundamental study on cellulase expression at genome-wide scale.
Figures

References
-
- Lin Y, Tanaka S. Ethanol fermentation from biomass resources: current state and prospects. Appl Microbiol Biotechnol. 2006;69:627–642. - PubMed
-
- Rouvinen J, Bergsfors T, Teeri T, Knowles KJC. Three-dimensional structure of cellobiohydrolase II from Trichoderma reesei. Science. 1990;249:380–386. - PubMed
-
- Divine C, Stahlberg J, Reinikaninen T, Rouhonen L, Petterson G, Knowles KJC, Teeri T, Jones TA. The three-dimensional crystal structure of the catalytic core of cellobiohydrolase I from Trichoderma reesei. Science. 1994;265:524–528. - PubMed
-
- Sandgren M, Stahlberg J, Mitchinson C. Structural and biochemical studies of GH family 12 cellulases: improved thermal stability, and ligand complexes. Prog Biophys Mol Biol. 2005;89:246–291. - PubMed
-
- Rojas AL, Fischer H, Eneiskaya EV, Kulminskaya AA, Shabalin KA, Neustroev KN, Craievich AF, Golubev AM, Polikarpov I. Structural insights into the beta-xylosidase from Trichoderma reesei obtained by synchrotron small-angle X-ray scattering and circular dichroism spectroscopy. Biochemistry. 2005;44:15578–15584. - PubMed
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

