Deubiquitination of CXCR4 by USP14 is critical for both CXCL12-induced CXCR4 degradation and chemotaxis but not ERK ativation
- PMID: 19106094
- PMCID: PMC2645827
- DOI: 10.1074/jbc.M808507200
Deubiquitination of CXCR4 by USP14 is critical for both CXCL12-induced CXCR4 degradation and chemotaxis but not ERK ativation
Abstract
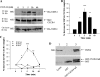
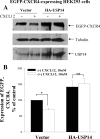
The chemokine receptor CXCR4 plays important roles in the immune and nervous systems. Abnormal expression of CXCR4 contributes to cancer and inflammatory and neurodegenerative disorders. Although ligand-dependent CXCR4 ubiquitination is known to accelerate CXCR4 degradation, little is known about counter mechanisms for receptor deubiquitination. CXCL12, a CXCR4 agonist, induces a time-dependent association of USP14 with CXCR4, or its C terminus, that is not mimicked by USP2A, USP4, or USP7, other members of the deubiquitination catalytic family. Co-localization of CXCR4 and USP14 also is time-dependent following CXCL12 stimulation. The physical interaction of CXCR4 and USP14 is paralleled by USP14-catalyzed deubiquitination of the receptor; knockdown of endogenous USP14 by RNA interference (RNAi) blocks CXCR4 deubiquitination, whereas overexpression of USP14 promotes CXCR4 deubiquitination. We also observed that ubiquitination of CXCR4 facilitated receptor degradation, whereas overexpression of USP14 or RNAi-induced knockdown of USP14 blocked CXCL12-mediated CXCR4 degradation. Most interestingly, CXCR4-mediated chemotactic cell migration was blocked by either overexpression or RNAi-mediated knockdown of USP14, implying that a CXCR4-ubiquitin cycle on the receptor, rather than a particular ubiquitinated state of the receptor, is critical for the ligand gradient sensing and directed motility required for chemokine-mediated chemotaxis. Our observation that a mutant of CXCR4, HA-3K/R CXCR4, which cannot be ubiquitinated and does not mediate a chemotactic response to CXCL12, indicates the importance of this covalent modification not only in marking receptors for degradation but also for permitting CXCR4-mediated signaling. Finally, the indistinguishable activation of ERK by wild typeor 3K/R-CXCR4 suggests that chemotaxis in response to CXCL12 may be independent of the ERK cascade.
Figures



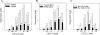
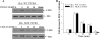
References
-
- Haribabu, B., Richardson, R. M., Fisher, I., Sozzani, S., Peiper, S. C., Horuk, R., Ali, H., and Snyderman, R. (1997) J. Biol. Chem. 272 28726-28731 - PubMed
-
- Balabanian, K., Lagane, B., Infantino, S., Chow, K. Y., Harriague, J., Moepps, B., Arenzana-Seisdedos, F., Thelen, M., and Bachelerie, F. (2005) J. Biol. Chem. 280 35760-35766 - PubMed
-
- Nagasawa, T., Hirota, S., Tachibana, K., Takakura, N., Nishikawa, S., Kitamura, Y., Yoshida, N., Kikutani, H., and Kishimoto, T. (1996) Nature 382 635-638 - PubMed
-
- D'Apuzzo, M., Rolink, A., Loetscher, M., Hoxie, J. A., Clark-Lewis, I., Melchers, F., Baggiolini, M., and Moser, B. (1997) Eur. J. Immunol. 27 1788-1793 - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Molecular Biology Databases
Miscellaneous

