Conformational variants of duplex DNA correlated with cytosine-rich chromosomal fragile sites
- PMID: 19106104
- PMCID: PMC2652318
- DOI: 10.1074/jbc.M806866200
Conformational variants of duplex DNA correlated with cytosine-rich chromosomal fragile sites
Abstract
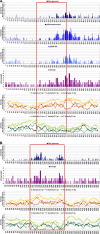
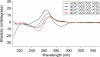
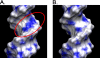
We found that several major chromosomal fragile sites in human lymphomas, including the bcl-2 major breakpoint region, bcl-1 major translocation cluster, and c-Myc exon 1-intron 1 boundary, contain distinctive sequences of consecutive cytosines exhibiting a high degree of reactivity with the structure-specific chemical probe bisulfite. To assess the inherent structural variability of duplex DNA in these regions and to determine the range of structures reactive to bisulfite, we have performed bisulfite probing on genomic DNA in vitro and in situ; on duplex DNA in supercoiled and linearized plasmids; and on oligonucleotide DNA/DNA and DNA/2'-O-methyl RNA duplexes. Bisulfite is significantly more reactive at the frayed ends of DNA duplexes, which is expected given that bisulfite is an established probe of single-stranded DNA. We observed that bisulfite also distinguishes between more subtle sequence/structural differences in duplex DNA. Supercoiled plasmids are more reactive than linear DNA; and sequences containing consecutive cytosines, namely GGGCCC, are more reactive than those with alternating guanine and cytosine, namely GCGCGC. Circular dichroism and x-ray crystallography show that the GGGCCC sequence forms an intermediate B/A structure. Molecular dynamics simulations also predict an intermediate B/A structure for this sequence, and probe calculations suggest greater bisulfite accessibility of cytosine bases in the intermediate B/A structure over canonical B- or A-form DNA. Electrostatic calculations reveal that consecutive cytosine bases create electropositive patches in the major groove, predicting enhanced localization of the bisulfite anion at homo-C tracts over alternating G/C sequences. These characteristics of homo-C tracts in duplex DNA may be associated with DNA-protein interactions in vivo that predispose certain genomic regions to chromosomal fragility.
Figures




Similar articles
-
Evidence for a triplex DNA conformation at the bcl-2 major breakpoint region of the t(14;18) translocation.J Biol Chem. 2005 Jun 17;280(24):22749-60. doi: 10.1074/jbc.M502952200. Epub 2005 Apr 18. J Biol Chem. 2005. PMID: 15840562
-
Nickel-guanine interactions in DNA: crystal structure of nickel-d[CGTGTACACG]2.J Biol Inorg Chem. 2002 Jan;7(1-2):195-9. doi: 10.1007/s007750100286. Epub 2001 Aug 25. J Biol Inorg Chem. 2002. PMID: 11862555
-
HhaI methyltransferase flips its target base out of the DNA helix.Cell. 1994 Jan 28;76(2):357-69. doi: 10.1016/0092-8674(94)90342-5. Cell. 1994. PMID: 8293469
-
The bisulfite reaction with cytosine and genomic DNA structure.Anal Biochem. 2024 Aug;691:115532. doi: 10.1016/j.ab.2024.115532. Epub 2024 Apr 10. Anal Biochem. 2024. PMID: 38609028 Review.
-
DNA structures at chromosomal translocation sites.Bioessays. 2006 May;28(5):480-94. doi: 10.1002/bies.20353. Bioessays. 2006. PMID: 16615081 Review.
Cited by
-
RNA Polymerase Collision versus DNA Structural Distortion: Twists and Turns Can Cause Break Failure.Mol Cell. 2016 May 5;62(3):327-334. doi: 10.1016/j.molcel.2016.03.034. Mol Cell. 2016. PMID: 27153532 Free PMC article. Review.
-
The mechanisms of human lymphoid chromosomal translocations and their medical relevance.Crit Rev Biochem Mol Biol. 2022 Jun;57(3):227-243. doi: 10.1080/10409238.2021.2004576. Epub 2021 Dec 7. Crit Rev Biochem Mol Biol. 2022. PMID: 34875186 Free PMC article.
-
Diversity upon diversity: linking DNA double-strand break repair to blood cancer health disparities.Trends Cancer. 2022 Apr;8(4):328-343. doi: 10.1016/j.trecan.2022.01.003. Epub 2022 Jan 31. Trends Cancer. 2022. PMID: 35094960 Free PMC article. Review.
-
Concerted dynamics of metallo-base pairs in an A/B-form helical transition.Nat Commun. 2019 Oct 23;10(1):4818. doi: 10.1038/s41467-019-12440-x. Nat Commun. 2019. PMID: 31645548 Free PMC article.
-
How does DNA break during chromosomal translocations?Nucleic Acids Res. 2011 Aug;39(14):5813-25. doi: 10.1093/nar/gkr223. Epub 2011 Apr 15. Nucleic Acids Res. 2011. PMID: 21498543 Free PMC article. Review.
References
-
- Saenger, W. (1984) Principles of Nucleic Acid Structure, Springer-Verlag, New York
-
- Calladine, C. R., Drew, H. R., Luisi, B. F., and Travers, A. A. (2004) Understanding DNA: The Molecule and How It Works, 3rd Ed., Elsevier/Academic Press, San Diego
-
- Sinden, R. R. (1994) DNA Structure and Function, Academic Press, San Diego
-
- Mirkin, S. M. (2006) Curr. Opin. Struct. Biol. 16 351-358 - PubMed
-
- Hud, N. V., and Plavec, J. (2003) Biopolymers 69 144-158 - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Other Literature Sources
Medical
Research Materials
Miscellaneous

