Cluster analysis and comparison of various chloroplast transcriptomes and genes in Arabidopsis thaliana
- PMID: 19106172
- PMCID: PMC2646353
- DOI: 10.1093/dnares/dsn031
Cluster analysis and comparison of various chloroplast transcriptomes and genes in Arabidopsis thaliana
Abstract
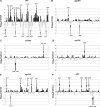
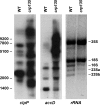
Chloroplast RNA metabolism is integrated into wider gene regulatory networks. To explore how, we performed a chloroplast genome-wide expression analysis on numerous nuclear Arabidopsis mutants affected in diverse chloroplast functions and wild-type plants subjected to various stresses and conditions. On the basis of clustering analysis, plastid genes could be divided into two oppositely regulated clusters, largely congruent with known targets of nucleus- and plastid-encoded RNA polymerases, respectively. Further eight sub-clusters contained co-transcribed and functionally tightly associated genes. The chloroplast transcriptomes could also be classified into two major groups comprising mutants preferentially affected in general plastid gene expression and other chloroplast functions, respectively. Deviations from characteristic expression profiles of transcriptomes served to identify novel mutants impaired in accumulation and/or processing of specific plastid RNAs. Expression profiles were useful to distinguish albino mutants affected in plastid gene expression from those with defects in other plastid functions. Remarkably, biotic and abiotic stressors did not define transcriptionally determined clusters indicating that post-transcriptional regulation of plastid gene expression becomes more important under changing environmental conditions. Overall, the identification of sets of co-regulated genes provides insights into the integration of plastid gene expression into common pathways that ensures a coordinated response.
Figures



References
-
- Barkan A., Goldschmidt-Clermont M. Participation of nuclear genes in chloroplast gene expression. Biochimie. 2000;82:559–572. - PubMed
-
- Nakamura T., Schuster G., Sugiura M., Sugita M. Chloroplast RNA-binding and pentatricopeptide repeat proteins. Biochem. Soc. Trans. 2004;32:571–574. - PubMed
-
- Tanaka K., Oikawa K., Ohta N., Kuroiwa H., Kuroiwa T., Takahashi H. Nuclear encoding of a chloroplast RNA polymerase sigma subunit in a red alga. Science. 1996;272:1932–1935. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources

