A comprehensive collection of experimentally validated primers for Polymerase Chain Reaction quantitation of murine transcript abundance
- PMID: 19108745
- PMCID: PMC2631021
- DOI: 10.1186/1471-2164-9-633
A comprehensive collection of experimentally validated primers for Polymerase Chain Reaction quantitation of murine transcript abundance
Abstract
Background: Quantitative polymerase chain reaction (QPCR) is a widely applied analytical method for the accurate determination of transcript abundance. Primers for QPCR have been designed on a genomic scale but non-specific amplification of non-target genes has frequently been a problem. Although several online databases have been created for the storage and retrieval of experimentally validated primers, only a few thousand primer pairs are currently present in existing databases and the primers are not designed for use under a common PCR thermal profile.
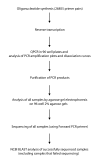
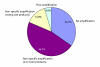
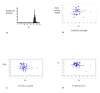
Results: We previously reported the implementation of an algorithm to predict PCR primers for most known human and mouse genes. We now report the use of that resource to identify 17483 pairs of primers that have been experimentally verified to amplify unique sequences corresponding to distinct murine transcripts. The primer pairs have been validated by gel electrophoresis, DNA sequence analysis and thermal denaturation profile. In addition to the validation studies, we have determined the uniformity of amplification using the primers and the technical reproducibility of the QPCR reaction using the popular and inexpensive SYBR Green I detection method.
Conclusion: We have identified an experimentally validated collection of murine primer pairs for PCR and QPCR which can be used under a common PCR thermal profile, allowing the evaluation of transcript abundance of a large number of genes in parallel. This feature is increasingly attractive for confirming and/or making more precise data trends observed from experiments performed with DNA microarrays.
Figures

References
-
- Bustin SA. A-Z of Quantitative PCR. San Diego: IUL Press; 2004.
-
- Walker NJ. A technique whose time has come. Science. 2002;296:557–559. - PubMed
-
- Schinke-Braun M, Couget JA. Expression profiling using affymetrix genechip probe arrays. Methods Mol Biol. 2007;366:13–40. - PubMed
-
- Higuchi R, Dollinger G, Walsh PS, Griffith R. Simultaneous amplification and detection of specific DNA sequences. Biotechnology (N Y) 1992;10:413–417. - PubMed
-
- Higuchi R, Fockler C, Dollinger G, Watson R. Kinetic PCR analysis: real-time monitoring of DNA amplification reactions. Biotechnology (N Y) 1993;11:1026–1030. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

