Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif
- PMID: 19109396
- PMCID: PMC2643725
- DOI: 10.1128/JVI.01898-08
Regulation of APOBEC3 proteins by a novel YXXL motif in human immunodeficiency virus type 1 Vif and simian immunodeficiency virus SIVagm Vif
Abstract
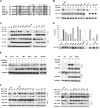
The APOBEC3 cytidine deaminases are potent antiviral factors that restrict the replication of human immunodeficiency virus type 1 (HIV-1). In HIV-1-infected CD4+ T cells, the viral accessory protein Vif binds to APOBEC3G (A3G), APOBEC3F (A3F), and APOBEC3C (A3C) and targets these proteins for polyubiquitination by forming an E3 ubiquitin ligase with cullin 5. Previous studies identified regions of HIV-1 Vif, 40YRHHY44 and 12QVDRMR17, which are important for interaction with A3G and A3F, respectively, and showed that Vif residues 54 to 71 are sufficient for A3G binding. Here, we identify 69YXXL72 as a novel conserved motif in HIV-1 Vif that mediates binding to human A3G and its subsequent degradation. Studies on other APOBEC3 proteins revealed that Tyr69 and Leu72 are important for the degradation of A3F and A3C as well. Similar to A3F, A3C regulation is also mediated by Vif residues 12QVDRMR17. Simian immunodeficiency virus (SIV) Vif was shown to bind and degrade African green monkey A3G (agmA3G) and, unexpectedly, human A3C. The YXXL motif of SIVagm Vif was important for the inactivation of agmA3G and human A3C. Unlike HIV-1 Vif, however, SIVagm Vif does not require Tyr40 and His43 for agmA3G degradation. Tyr69 in the YXXL motif was critical for binding of recombinant glutathione S-transferase-Vif(1-94) to A3G in vitro. These results suggest that the YXXL motif in Vif is a potential target for small-molecule inhibitors to block Vif interaction with A3G, A3F, and A3C, and thereby protect cells against HIV-1 infection.
Figures
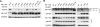
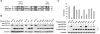
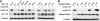

References
-
- Chiu, Y. L., V. B. Soros, J. F. Kreisberg, K. Stopak, W. Yonemoto, and W. C. Greene. 2005. Cellular APOBEC3G restricts HIV-1 infection in resting CD4+ T cells. Nature 435108-114. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Research Materials

