Accurate SHAPE-directed RNA structure determination
- PMID: 19109441
- PMCID: PMC2629221
- DOI: 10.1073/pnas.0806929106
Accurate SHAPE-directed RNA structure determination
Abstract
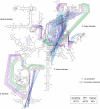
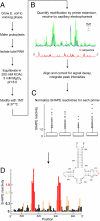
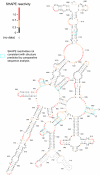
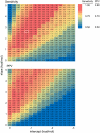
Almost all RNAs can fold to form extensive base-paired secondary structures. Many of these structures then modulate numerous fundamental elements of gene expression. Deducing these structure-function relationships requires that it be possible to predict RNA secondary structures accurately. However, RNA secondary structure prediction for large RNAs, such that a single predicted structure for a single sequence reliably represents the correct structure, has remained an unsolved problem. Here, we demonstrate that quantitative, nucleotide-resolution information from a SHAPE experiment can be interpreted as a pseudo-free energy change term and used to determine RNA secondary structure with high accuracy. Free energy minimization, by using SHAPE pseudo-free energies, in conjunction with nearest neighbor parameters, predicts the secondary structure of deproteinized Escherichia coli 16S rRNA (>1,300 nt) and a set of smaller RNAs (75-155 nt) with accuracies of up to 96-100%, which are comparable to the best accuracies achievable by comparative sequence analysis.
Conflict of interest statement
The authors declare no conflict of interest.
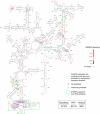
Figures
References
-
- Buchmueller KL, Webb AE, Richardson DA, Weeks KM. A collapsed, non-native RNA folding state. Nat Struct Biol. 2000;7:362–366. - PubMed
-
- Gesteland RF, Cech TR, Atkins JF. The RNA World. 3rd Ed. Cold Spring Harbor, NY: Cold Spring Harbor Laboratory Press; 2006.
-
- Kozak M. Regulation of translation via mRNA structure in prokaryotes and eukaryotes. Gene. 2005;361:13–37. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources
Research Materials

