The pathogen receptor liver and lymph node sinusoidal endotelial cell C-type lectin is expressed in human Kupffer cells and regulated by PU.1
- PMID: 19111020
- PMCID: PMC7165556
- DOI: 10.1002/hep.22678
The pathogen receptor liver and lymph node sinusoidal endotelial cell C-type lectin is expressed in human Kupffer cells and regulated by PU.1
Abstract
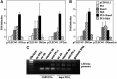
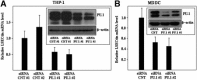
Human LSECtin (liver and lymph node sinusoidal endothelial cell C-type lectin, CLEC4G) is a C-type lectin encoded within the L-SIGN/DC-SIGN/CD23 gene cluster. LSECtin acts as a pathogen attachment factor for Ebolavirus and the SARS coronavirus, and its expression can be induced by interleukin-4 on monocytes and macrophages. Although reported as a liver and lymph node sinusoidal endothelial cell-specific molecule, LSECtin could be detected in the MUTZ-3 dendritic-like cell line at the messenger RNA (mRNA) and protein level, and immunohistochemistry analysis on human liver revealed its presence in Kupffer cells coexpressing the myeloid marker CD68. The expression of LSECtin in myeloid cells was further corroborated through the analysis of the proximal regulatory region of the human LSECtin gene, whose activity was maximal in LSECtin+ myeloid cells, and which contains a highly conserved PU.1-binding site. PU.1 transactivated the LSECtin regulatory region in collaboration with hematopoietic-restricted transcription factors (Myb, RUNX3), and was found to bind constitutively to the LSECtin proximal promoter. Moreover, knockdown of PU.1 through the use of small interfering RNA led to a decrease in LSECtin mRNA levels in THP-1 and monocyte-derived dendritic cells, thus confirming the involvement of PU.1 in the myeloid expression of the lectin.
Conclusion: LSECtin is expressed by liver myeloid cells, and its expression is dependent on the PU.1 transcription factor.
Figures





References
-
- Geijtenbeek TB, Torensma R, van Vliet SJ, van Duijnhoven GC, Adema GJ, van Kooyk Y, et al. Identification of DC‐SIGN, a novel dendritic cell‐specific ICAM‐3 receptor that supports primary immune responses. Cell 2000; 100: 575–585. - PubMed
-
- Soilleux EJ, Barten R, Trowsdale J. DC‐SIGN; a related gene, DC‐SIGNR; and CD23 form a cluster on 19p13. J Immunol 2000; 165: 2937–2942. - PubMed
-
- Bashirova AA, Geijtenbeek TB, van Duijnhoven GC, van Vliet SJ, Eilering JB, Martin MP, et al. A dendritic cell‐specific intercellular adhesion molecule 3‐grabbing nonintegrin (DC‐SIGN)‐related protein is highly expressed on human liver sinusoidal endothelial cells and promotes HIV‐1 infection. J Exp Med 2001; 193: 671–678. - PMC - PubMed
-
- Liu W, Tang L, Zhang G, Wei H, Cui Y, Guo L, et al. Characterization of a novel C‐type lectin‐like gene, LSECtin: demonstration of carbohydrate binding and expression in sinusoidal endothelial cells of liver and lymph node. J Biol Chem 2004; 279: 18748–18758. - PubMed
-
- Dominguez‐Soto A, Aragoneses‐Fenoll L, Martin‐Gayo E, Martinez‐Prats L, Colmenares M, Naranjo‐Gomez M, et al. The DC‐SIGN‐related lectin LSECtin mediates antigen capture and pathogen binding by human myeloid cells. Blood 2007; 109: 5337–5345. - PubMed
Publication types
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
