Chromosomal inversions between human and chimpanzee lineages caused by retrotransposons
- PMID: 19112500
- PMCID: PMC2603318
- DOI: 10.1371/journal.pone.0004047
Chromosomal inversions between human and chimpanzee lineages caused by retrotransposons
Abstract
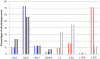
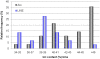
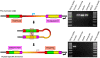
The long interspersed element-1 (LINE-1 or L1) and Alu elements are the most abundant mobile elements comprising 21% and 11% of the human genome, respectively. Since the divergence of human and chimpanzee lineages, these elements have vigorously created chromosomal rearrangements causing genomic difference between humans and chimpanzees by either increasing or decreasing the size of genome. Here, we report an exotic mechanism, retrotransposon recombination-mediated inversion (RRMI), that usually does not alter the amount of genomic material present. Through the comparison of the human and chimpanzee draft genome sequences, we identified 252 inversions whose respective inversion junctions can clearly be characterized. Our results suggest that L1 and Alu elements cause chromosomal inversions by either forming a secondary structure or providing a fragile site for double-strand breaks. The detailed analysis of the inversion breakpoints showed that L1 and Alu elements are responsible for at least 44% of the 252 inversion loci between human and chimpanzee lineages, including 49 RRMI loci. Among them, three RRMI loci inverted exonic regions in known genes, which implicates this mechanism in generating the genomic and phenotypic differences between human and chimpanzee lineages. This study is the first comprehensive analysis of mobile element bases inversion breakpoints between human and chimpanzee lineages, and highlights their role in primate genome evolution.
Conflict of interest statement
Figures

References
-
- Deininger PL, Moran JV, Batzer MA, Kazazian HH., Jr Mobile elements and mammalian genome evolution. Curr Opin Genet Dev. 2003;13:651–658. - PubMed
-
- Luan DD, Korman MH, Jakubczak JL, Eickbush TH. Reverse transcription of R2Bm RNA is primed by a nick at the chromosomal target site: a mechanism for non-LTR retrotransposition. Cell. 1993;72:595–605. - PubMed
-
- Mathias SL, Scott AF, Kazazian HH, Jr, Boeke JD, Gabriel A. Reverse transcriptase encoded by a human transposable element. Science. 1991;254:1808–1810. - PubMed
-
- Deininger PL, Batzer MA, Hutchison CA, 3rd, Edgell MH. Master genes in mammalian repetitive DNA amplification. Trends Genet. 1992;8:307–311. - PubMed
-
- Weiner AM. Do all SINEs lead to LINEs? Nat Genet. 2000;24:332–333. - PubMed
Publication types
MeSH terms
Substances
Grants and funding
LinkOut - more resources
Full Text Sources
Other Literature Sources

