Identification of two novel OPA1 mutations in Chinese families with autosomal dominant optic atrophy
- PMID: 19112530
- PMCID: PMC2610289
Identification of two novel OPA1 mutations in Chinese families with autosomal dominant optic atrophy
Abstract
Purpose: To report the clinical features and identification of two novel mutations in two Chinese pedigrees with autosomal dominant optic atrophy (ADOA).
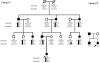
Methods: Two families (F1 and F2) including ten affected members and nine unaffected family individuals were examined clinically. After informed consent was obtained, peripheral blood samples of all the participants were obtained, and genomic DNA was extracted. Linkage analysis was performed with two microsatellite markers around the OPA1 gene (D3S2305 and D3S3562) in family F1. The coding region (exon 1-28), including intron-exon boundary of the OPA1 gene, were screened in the 2 families by polymerase chain reaction (PCR) and direct DNA sequencing. Whenever substitutions were identified in a patient, single strand conformation polymorphism (SSCP) analysis was performed on all available family members and 100 normal controls. To characterize a splicing site mutation, RT-PCR of total RNA of leukocytes obtained from three patients and seven unaffected individuals of family F1 was performed with the specific primers.
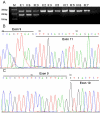
Results: The affected individuals all presented with bilateral visual failure and temporal or total pallor of the optic discs. Genotyping of family F1 revealed the linkage to the OPA1 gene on 3q28-29. After sequencing of OPA1 gene, a novel heterozygous splicing site mutation c.985 -2A>G in intron 9 was found in family F1. RT-PCR result showed the skipping of the exon 10 in the mutant transcript, which results in loss of 27 amino acids in the OPA1 protein. A novel heterozygous nonsense mutation c.2197C>T(p.R733X)was detected in family F2.
Conclusions: Our findings expand the spectrum of OPA1 mutations and further established the role of OPA1 gene in Chinese patients with ADOA.
Figures

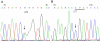
Similar articles
-
First report of OPA1 screening in Greek patients with autosomal dominant optic atrophy and identification of a previously undescribed OPA1 mutation.Mol Vis. 2014 May 27;20:691-703. eCollection 2014. Mol Vis. 2014. PMID: 24883014 Free PMC article.
-
Novel mutations of the OPA1 gene in Chinese dominant optic atrophy.Ophthalmology. 2010 Feb;117(2):392-6.e1. doi: 10.1016/j.ophtha.2009.07.019. Epub 2009 Dec 6. Ophthalmology. 2010. PMID: 19969356
-
A comprehensive survey of mutations in the OPA1 gene in patients with autosomal dominant optic atrophy.Invest Ophthalmol Vis Sci. 2002 Jun;43(6):1715-24. Invest Ophthalmol Vis Sci. 2002. PMID: 12036970
-
Meta-analysis of genotype-phenotype analysis of OPA1 mutations in autosomal dominant optic atrophy.Mitochondrion. 2019 May;46:262-269. doi: 10.1016/j.mito.2018.07.006. Epub 2018 Aug 27. Mitochondrion. 2019. PMID: 30165240
-
Recessive optic atrophy, sensorimotor neuropathy and cataract associated with novel compound heterozygous mutations in OPA1.Mol Med Rep. 2016 Jul;14(1):33-40. doi: 10.3892/mmr.2016.5209. Epub 2016 May 4. Mol Med Rep. 2016. PMID: 27150940 Free PMC article. Review.
Cited by
-
First report of OPA1 screening in Greek patients with autosomal dominant optic atrophy and identification of a previously undescribed OPA1 mutation.Mol Vis. 2014 May 27;20:691-703. eCollection 2014. Mol Vis. 2014. PMID: 24883014 Free PMC article.
-
Mitochondrial Mutations in Ethambutol-Induced Optic Neuropathy.Front Cell Dev Biol. 2021 Oct 5;9:754676. doi: 10.3389/fcell.2021.754676. eCollection 2021. Front Cell Dev Biol. 2021. PMID: 34676220 Free PMC article.
-
Solving a 50 year mystery of a missing OPA1 mutation: more insights from the first family diagnosed with autosomal dominant optic atrophy.Mol Neurodegener. 2010 Jun 14;5:25. doi: 10.1186/1750-1326-5-25. Mol Neurodegener. 2010. PMID: 20546606 Free PMC article.
-
Optic atrophy plus phenotype due to mutations in the OPA1 gene: two more Italian families.J Neurol Sci. 2012 Apr 15;315(1-2):146-9. doi: 10.1016/j.jns.2011.12.002. Epub 2011 Dec 22. J Neurol Sci. 2012. PMID: 22197506 Free PMC article.
-
Processing of OPA1 with a novel N-terminal mutation in patients with autosomal dominant optic atrophy: Escape from nonsense-mediated decay.PLoS One. 2017 Aug 25;12(8):e0183866. doi: 10.1371/journal.pone.0183866. eCollection 2017. PLoS One. 2017. PMID: 28841713 Free PMC article.
References
-
- Kjer B, Eiberg H, Kjer P, Rosenberg T. Dominant optic atrophy mapped to chromosome 3q region. II. Clinical and epidemiological aspects. Acta Ophthalmol Scand. 1996;74:3–7. - PubMed
-
- Lyle WM. Genetic risks: a reference for eye care practitioners. Waterloo (Canada): University of Waterloo Press; 1990.
-
- Johnston PB, Gaster RN, Smith VC, Tripathi R. A clinicopathologic study of autosomal dominant optic atrophy. Am J Ophthalmol. 1979;88:868–75. - PubMed
-
- Eiberg H, Kjer B, Kjer P, Rosenberg T. Dominant optic atrophy (OPA1) mapped to chromosome 3q region I. Linkage analysis. Hum Mol Genet. 1994;3:977–80. - PubMed
MeSH terms
Substances
LinkOut - more resources
Full Text Sources
Miscellaneous
